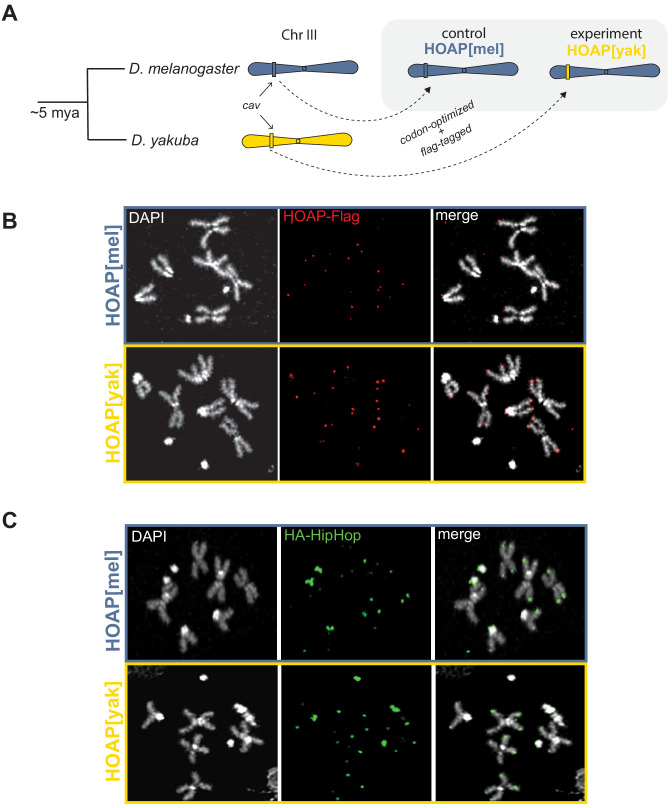
Figure 1. Allele swap strategy and phenotypic rescue by a diverged version of HOAP.
(A) Using CRISPR/Cas9-mediated transgenesis, we replaced the native coding sequence of cav/HOAP with either a Flag-tagged D. melanogaster coding sequence or instead a Flag-tagged D. yakuba coding sequence. Both coding sequences were intron-less and codon-optimized for D. melanogaster. (B) Mitotic chromosome squashes from larval brains homozygous for HOAP[mel]-Flag or HOAP[yak]-Flag stained with anti-Flag. (C) Mitotic chromosome squashes from larval brains homozygous for HA-HipHop, HOAP[mel]-Flag or HA-HipHop, HOAP[yak]-Flag stained with anti-HA.