Extended Data Fig. 1. PG modifications identified on proteobacterial OM proteins.
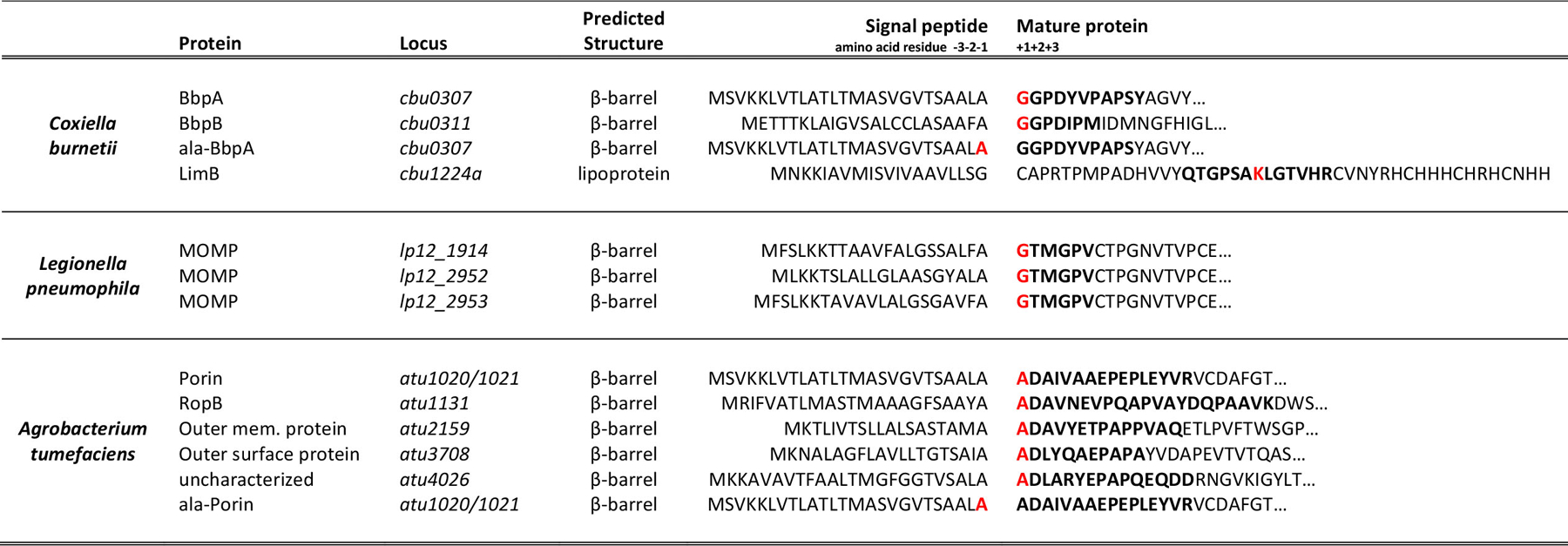
Proteins from A. tumefaciens, C. burnetii, and L. pneumophila are covalently attached to PG. Proteins identified in MS/MS data that contained a PG modification scoring higher than the first decoy and containing greater than 40% of the expected band y- ions were established as cut-off criteria. Predicted signal peptide cleavage sites of OM proteins covalently attached to PG from C. burnetii, L. pneumophila, and A. tumefaciens are shown. Amino acid numbering is based on the predicted signal peptide cleavage site. Peptide sequences that were found covalently attached to PG are bolded. Residuces with a PG tripeptide modification are also highlighted in red.
