Fig. 1. β-barrel OM proteins are covalently attached to PG and form a tight tether in C. burnetii.
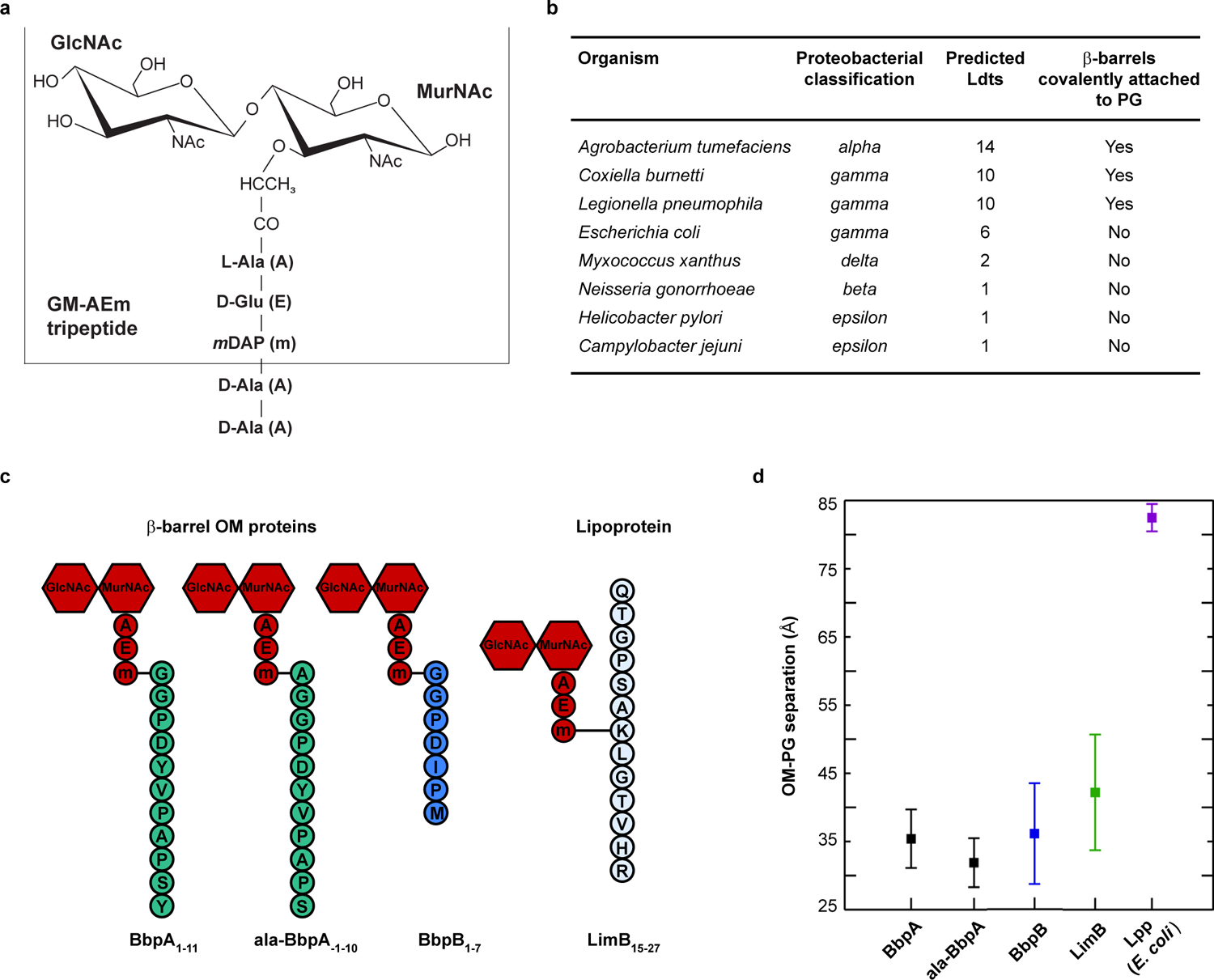
a. Schematic of the canonical Gram-negative muropeptide structure consisting of β−1–4 linked N-acetylglucosamine-N-acetylmuramic acid sugars [GlcNAc-MurNAc (GM)] attached to pentapeptide stems composed of L-alanine (A), γ-D-glutamic acid (E), meso-diaminopimelic acid (m), D-alanine (A), and D-alanine (A). The GM-AEm tripeptide moiety is outlined b. β-barrel OM proteins covalently attached to PG were identified in several proteobacteria. The number of predicted Ldts in each organism is shown. c. Schematic of the four covalently attached peptides identified in PG harvested from C. burnetii. Peptides are attached to the mDAP residue of the GM-AEm tripeptide moiety (red). d. Molecular dynamics simulation of C. burnetti OM protein-tethered models. The distance in angstroms between the centers-of-mass of the PG and the phosphorus atoms of the inner leaflet of the OM was measured for three independent 400 ns simulations of BbpA, ala-BbpA, BbpB, LimB, and E. coli Lpp. Distances are shown as averages with standard deviations.
