Extended Data Fig. 6. Extended Data Figure S6. Structural modeling of C. burnetii OM proteins.
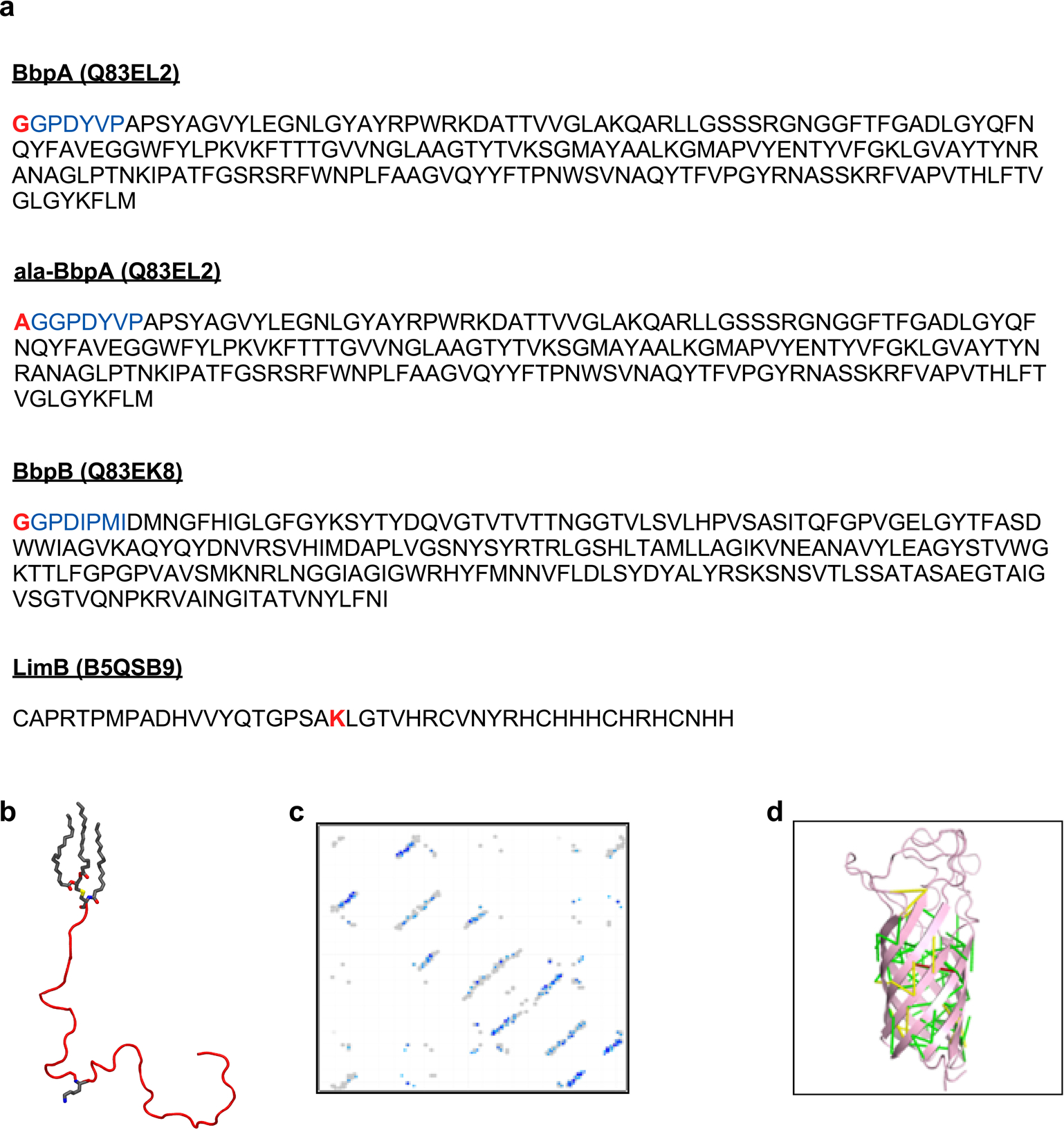
a. The protein sequences of BbpA, BbpB, and LimB are shown without N-terminal signal peptides. The disordered N-terminal domains of BbpA and BbpB were excluded from structural models but were included in subsequent molecular dynamics simulations (blue). Residues bound to PG in MD simulations are highlighted in red. b. The mature LimB was modeled as a random coil lacking any appreciable secondary structure. The three acyl tails at the N-terminus are shown in grey as is Lys21. c. Contact map predicted from coevolution analysis of BbpA. Contacts are shown in shades of blue (darker blue = higher probability) and contacts from E. coli OmpA (PDB entry 1QJP) are shown in gray. d. Predicted contacts mapped onto the final Rosetta model of BbpA. Contacts are color coded by Cα-Cα distance: green (<5 Å), yellow (5–10 Å), red (>10 Å).
