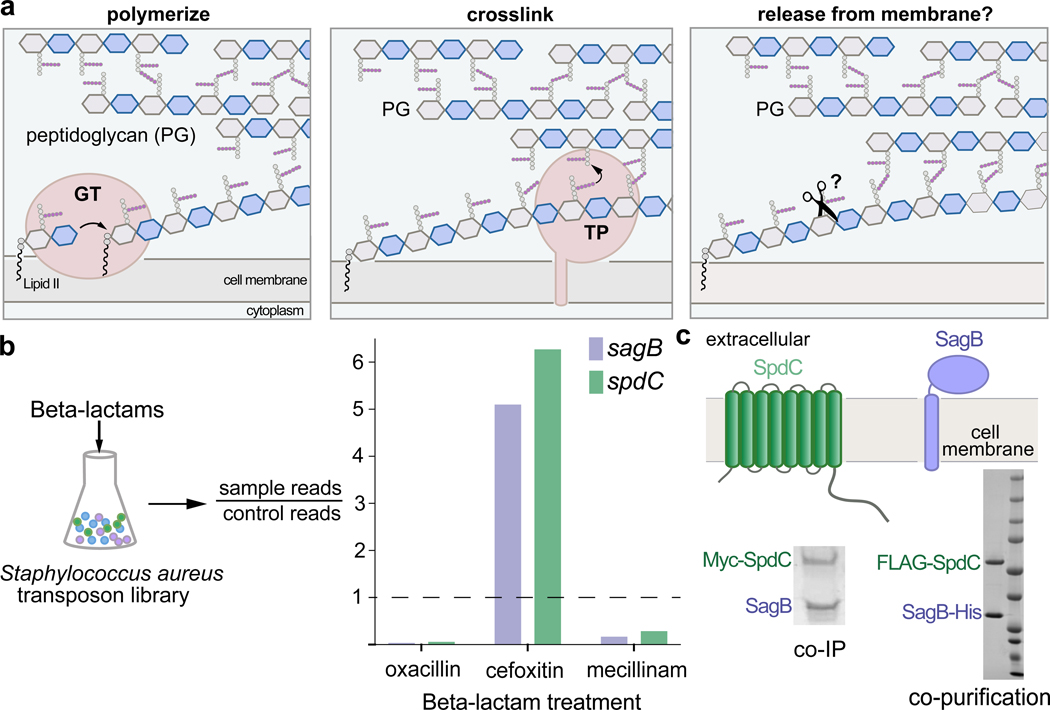
Figure 1. The cell wall hydrolase SagB and the membrane protein SpdC form a complex.
a, Overview of the final steps in peptidoglycan assembly. Left panel: After translocation to the outer face of the cytoplasmic membrane, the peptidoglycan (PG) monomer Lipid II is polymerized into linear glycan strands by glycosyltransferases (GT). Middle panel: Transpeptidase domains (TPs) crosslink glycan strands into the cell wall. Right panel: The glycan strand must be released from the membrane through some type of cleavage process in order to be incorporated into the cell wall. b, A Staphylococcus aureus transposon library made by combining six sublibraries as previously reported9,10 was treated with a panel of beta-lactams (oxacillin, cefoxitin, mecillinam) that have different selectivities for the four native S. aureus penicillin-binding proteins (PBPs)11. Only two genes, sagB and spdC, displayed a response pattern in which transposon reads were depleted under oxacillin and mecillinam treatment but enriched under cefoxitin treatment. Corrected p-values are reported for this experiment in Supplementary Table 1. c, SagB is a membrane-anchored glucosaminidase12,13 and SpdC is an eight-pass membrane protein15. Myc-tagged SpdC was expressed in a ΔspdC S. aureus strain and co-immunoprecipitated from solubilized membranes. (“Co-IP”, see also Supplementary Fig. 1). SagB was identified by LC-MS-MS analysis (Supplementary Table 1). Tandem affinity purification of SagB-His6 and FLAG-SpdC from E. coli yielded a stable 1:1 complex (“co-purification”, see also Supplementary Figure 2).