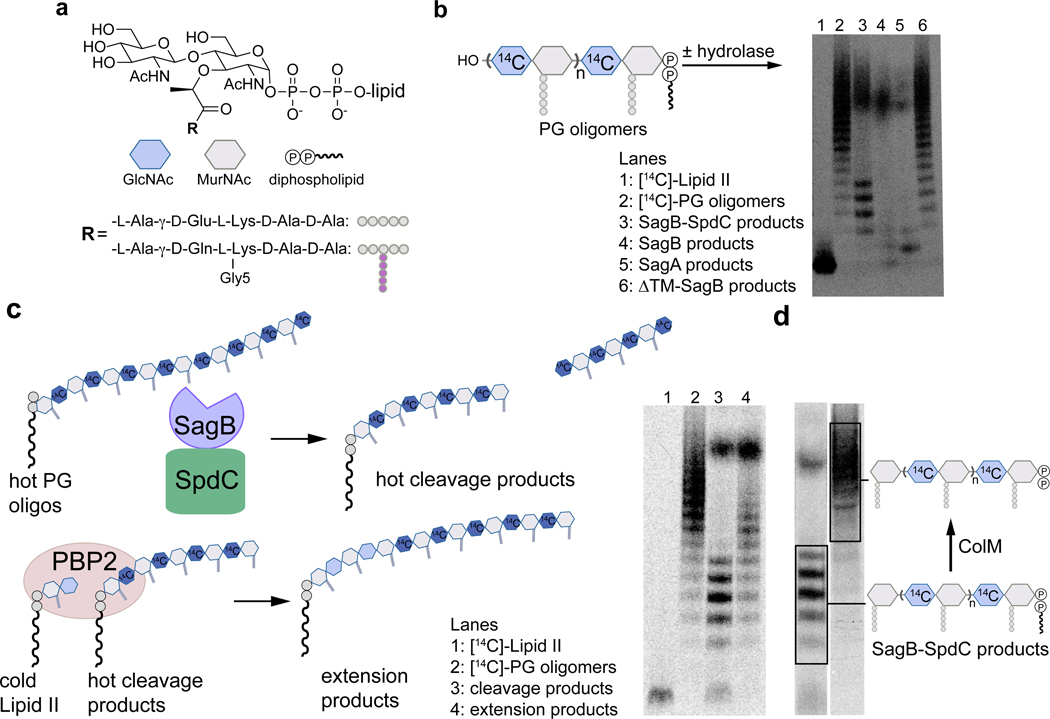
Figure 2. In vitro reconstitution shows that SagB-SpdC cleaves nascent peptidoglycan to short lipid-linked oligomers that can be elongated.
a, Chemical and cartoon representations of the synthetic Lipid II analog18 and native S. aureus Lipid II that were used to prepare peptidoglycan polymers in panels b-d. b, Radiolabeled peptidoglycan polymers were incubated with the SagB-SpdC complex, SagB alone, SagA, or SagB lacking its transmembrane helix. The signal towards the top of the autoradiograph in lanes 3–5 corresponds to short, lipid-free peptidoglycan fragments, but the distribution of lengths differs (Supplementary Figure 5). SagB-SpdC also produces a short ladder of radiolabeled peptidoglycan fragments (see also Supplementary Figure 4). c, Left: schematic of assay to determine whether SagB-SpdC product bands contained a lipid-anchor. Right: Radiolabeled SagB-SpdC products were incubated with unlabeled Lipid II and PBP2 and were extended to longer products. d, The bacteriocin Colicin M (ColM) de-lipidates Lipid II and peptidoglycan oligomers, but leaves the anomeric diphosphate (Supplementary Figures 6 and 7)23,24. Incubation of SagB-SpdC products (lane 3) with ColM (lane 4) resulted in the complete disappearance of fastmigrating bands and the appearance of slower-migrating products. Product characterization by LC-MS confirmed the indicated structure (Supplementary Figure 8). The faster migration of the SagB-SpdC products containing a lipid may be due to SDS binding to the lipid and increasing the net negative charge of these species. Experiments were performed at least three times in biological replicates.