Figure 6.
Schematic Illustration of and Data Mining of Non-Full-Length Receptors
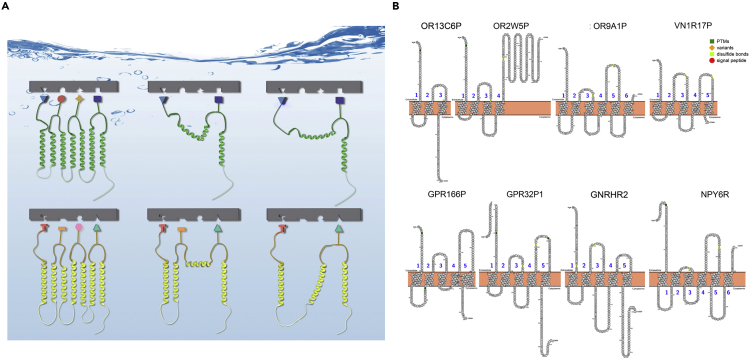
(A) Schematic illustration of possible ligand interaction of non-full-length CXCR4 and CCR5 receptors. The ligand-binding motif in N terminus and 3 EC loops are simplified and represented with cartoon blocks. Y2H screen indicated EC3 to be an essential part for CXCL12 binding by nfCXCR4QTY receptors SZ146a and SZ158a. The inter-connect coil between the N terminus and EC3 only slightly reduced the ligand affinity. For nfCCR5QTY, even though SZ218a contains ligand-binding motif EC2 loop, the inter-coil in between EC1 and EC3 may cause undesired spacing between the functional sites and rendered a slightly reduced affinity compared with EC3 containing SZ190b. In these cases, EC3 loop is required for both nfCXCR4QTY and nfCCR5QTY ligand binding as first identified in Y2H in vivo selections.
(B) Truncated or mutated GPCRs without full 7TM. Eight truncated GPCRs are mined from the genome database that include three olfactory receptors and one vomeronasal receptor, GNRHR2 (gonadotropin releasing hormone receptor 2), NPY6R (putative neuropeptide Y receptor type 6), and putative GPCRs of unknown function. Common GPCRs have 7TM, but these truncated GPCRs with various deletions have 3TM, 4TM, 5TM, and 6TM. They are presumed to be non-functional. However, no experiments have been carried out to test their biological function. It is plausible that some of them may be still able to bind their respective ligands and carry out signaling in cells.
See also Figure S9.