Figure 1.
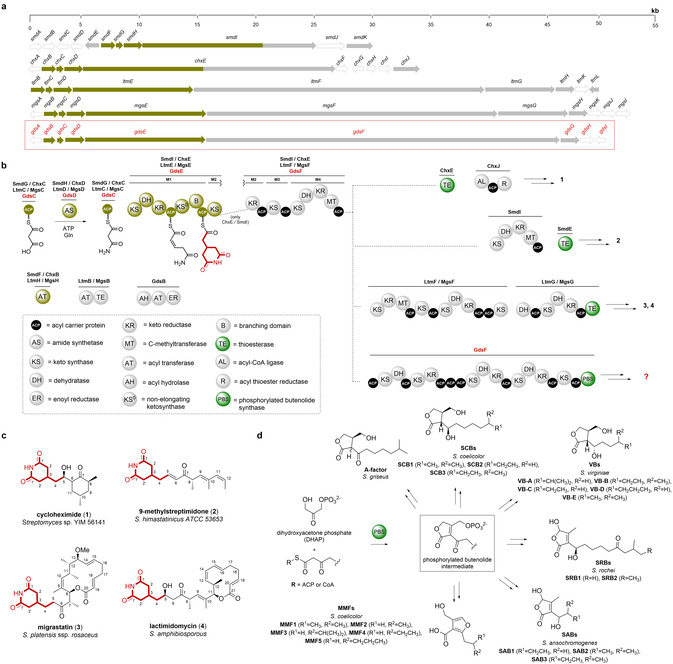
Comparison of Streptomyces glutarimide biosynthetic gene clusters and polyketide synthases they encode with that encoded by the cryptic B. gladioli gene cluster and role played by AfsA‐like phosphorylated butenolide synthases (PBSs) in Streptomyces signalling molecule biosynthesis. a) Comparison of biosynthetic gene clusters (BGCs) that direct glutarimide biosynthesis in Streptomyces species (9‐methylstreptimidone (smd), cycloheximide (chx), lactimidomycin (ltm) and migrastatin (mgs)) with the cryptic B. gladioli BCC0238/BCC1622 gene cluster (highlighted by the red box). The genes (or regions thereof) encoding the machinery responsible for assembling the common 2‐(2,6‐dioxopiperidin‐4‐yl)acetyl thioester biosynthetic intermediate are highlighted in gold. b) Comparison of the 9‐methylstreptimidone, cycloheximide, lactimidomycin and migrastatin PKS architectures with the PKS encoded by the cryptic B. gladioli BGC. Domains highlighted in dark yellow are involved in constructing the common 2‐(2,6‐dioxopiperidin‐4‐yl)acetyl thioester intermediate. Domains responsible for chain release (TE and PBS) are highlighted in green. Abbreviations for protein domains are displayed in the dashed box. c) Structures of glutarimide antibiotics produced by Streptomyces species. d) Structures of Streptomyces signalling molecules known or proposed to originate from a phosphorylated butenolide intermediate resulting from PBS‐catalysed condensation of DHAP with a 3‐ketoacyl‐ACP/CoA thioester.
