Abstract
Purpose
Chemical exchange saturation transfer MRI can provide accurate pH images, but the slow scan time (due to long saturation periods and multiple offsets sampling) reduce both the volume coverage and spatial resolution capability, hence the possibility to interrogate the heterogeneity in tumors and organs. To overcome these limitations, we propose a fast multislice CEST‐MRI sequence with high pH accuracy and spatial resolution.
Methods
The sequence first uses a long saturation pulse to induce the steady‐state CEST contrast and a second short saturation pulse repeated after each image acquisition to compensate for signal losses based on an uneven irradiation scheme combined with a single‐shot rapid acquisition with refocusing echoes readout. Sequence sensitivity and accuracy in measuring pH was optimized by simulation and assessed by in vitro studies in pH‐varying phantoms. In vivo validation was performed in two applications by acquiring multislice pH images covering the whole tumors and kidneys after iopamidol injection.
Results
Simulated and in vivo data showed comparable contrast efficiency and pH responsiveness by reducing saturation time. The experimental data from a homogeneous, pH‐varying, iopamidol‐containing phantom show that the sequence produced a uniform CEST contrast across slices and accurate values across slices in less than 10 minutes. In vivo measurements allowed us to quantify the 3D pH gradients of tumors and kidneys, with pH ranges comparable with the literature.
Conclusion
The proposed fast multislice CEST‐MRI sequence allows volumetric acquisitions with good pH sensitivity, accuracy, and spatial resolution for several in vivo pH imaging applications.
Keywords: 3D, CEST, extracellular pH, iopamidol, MRI, RARE
1. INTRODUCTION
The CEST mechanism is based on the selective saturation of low‐concentration endogenous or exogenous proton pools in exchange with water. During the exchange, saturated magnetization is transferred to water, and the resulting reduction in water signal allows the indirect detection of the CEST agent. 1 , 2 The CEST effect depends on different environmental and molecular factors, including labile proton concentration and exchange rate, and parameters that affect the exchange rate itself, such as pH and temperature. 3 , 4 , 5 Several studies have investigated the CEST technique in clinical applications such as oncology, 6 , 7 , 8 , 9 , 10 , 11 brain imaging, 12 , 13 , 14 , 15 measurements of metabolites in muscles 16 , 17 , 18 and glycosaminoglycans in knees and tendons, 19 , 20 and pH measurements in the bladder. 21 Among the multiple data that the CEST approach can provide, measurement of tissue pH has emerged as a potential marker of pathological conditions. 22 Iopamidol, an X‐ray contrast agent, has been used extensively to image tumor acidosis and perfusion, 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 to asses kidney pH evolution both in healthy and pathological conditions, 31 , 32 , 33 , 34 , 35 and for measuring cartilage pH. 36
However, long imaging times caused by multi‐offsets sampling and long saturation pulses limit the CEST‐MRI technique to single‐slice acquisitions that may not fully interrogate the spatial heterogeneity of tumors 37 or other organs, 27 thus limiting the functional information that can be provided. Therefore, fast volumetric readouts have been developed for the CEST‐MRI approaches to provide similar volume coverage as with other MRI‐based techniques. 38 At the clinical level, especially for brain imaging, CEST volumetric acquisition has been developed using both multislice 39 , 40 and 3D approaches. 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 Radiofrequency saturation has been accomplished using both continuous block pulses 40 , 42 , 51 and train of pulses, 25 , 39 , 41 , 43 , 46 , 47 , 48 , 49 , 52 which showed comparable saturation efficiency. Several fast acquisition schemes have been proposed: gradient echo (GRE), 39 , 40 , 41 , 45 , 47 , 48 , 49 turbo spin echo, 42 , 50 gradient spin echo–based approaches, 45 as well as Echo‐Planar Imaging (EPI) approaches, 43 , 46 both at low and high field. The GRAPPA technique, 46 , 47 , 49 compressed sensing, 42 SENSE, 43 , 45 , 50 , 51 keyhole, 53 and other acceleration strategies, including fingerprinting, 54 have been used to further reduce acquisition time. A great part of these approaches has been implemented by major MRI scanner providers with several advantages when compared with single‐slice acquisitions, but with potential limitations. Multislice acquisitions suffer from inherent contrast losses along the slices due to signal recovery, whereas 3D acquisitions often require dedicated hardware for parallel imaging to speed up the acquisition.
Conversely, at the preclinical level, only a few multislice CEST sequences have been investigated and implemented on preclinical MRI scanners to provide whole‐volume coverage. Conventionally, CEST imaging is implemented using a long irradiation period, followed by a fast readout scheme. Sun et al associated a long saturation period with an EPI readout and compensated for T1‐related signal losses along slices during the postprocessing. 55 Dixon et al, instead, described a GRE‐based acquisition scheme repeated after a very short saturation for the acquisition of every slice. 56 Combining the previously described approaches, Sun et al proposed an unevenly segmented saturation period and single‐shot EPI readout. 57 Moreover, a CEST fast imaging with steady‐state free precision (FISP) phase‐offset multiplanar (POMP) acquisition protocol that allows the simultaneous acquisition of several slices has been presented recently. 37 All previous approaches have been applied successfully in vivo, but some limitations are evident: the FISP and EPI readouts are prone to image artifacts and distortion, whereas the POMP technique suffers from phase‐encoding artifacts and is affected by B0 inhomogeneities. 58 Moreover, all approaches generally show a lower SNR compared with other fast readouts, such as those based on spin echo or fast spin echo. 59
In this work, we developed a new CEST sequence for rapid and volumetric acquisition to measure pH in organs. As previously done in Sun et al, 57 we propose a multislice acquisition based on the segmentation of the irradiation scheme in a long primary saturation period that introduces the steady‐state CEST contrast followed by a short secondary saturation period that recovers the saturation loss due to T1 and T2 relaxations before each slice acquisition. We exploited a single‐shot rapid acquisition with refocusing echoes (RARE) scheme, due to the expected higher SNR and lower artifacts. With the aim of reducing the acquisition time but still preserving a good CEST contrast, we investigated different saturation parameters and conditions. In addition, we compared the optimized proposed sequence with a single‐slice CEST‐MRI scheme, in terms of CEST contrast homogeneity along the slices. Furthermore, we validated the accuracy of the optimized sequence to measure pH in vitro and evaluated the capability of assessing in vivo pH heterogeneity in two representative tissues: kidneys and tumors.
2. THEORY
2.1. Sequence design
The two investigated sequence schemes based on a RARE readout are shown in Figure 1. Figure 1A shows a conventional single‐slice sequence, and Figure 1B shows the proposed multislice sequence. Single‐slice CEST sequence contains a long continuous‐wave irradiation block pulse of duration TS and amplitude B1, followed by a single‐shot RARE readout. The multislice CEST sequence combines the uneven saturation scheme described in Sun et al 57 with a RARE acquisition readout. The magnetization‐transfer module is unevenly segmented in two periods: a long primary RF irradiation block pulse of duration TS1, which generates the steady state, and multiple short repetitive secondary RF pulses of duration (TS2) applied immediately after each slice acquisition, which restore the saturation.
FIGURE 1.
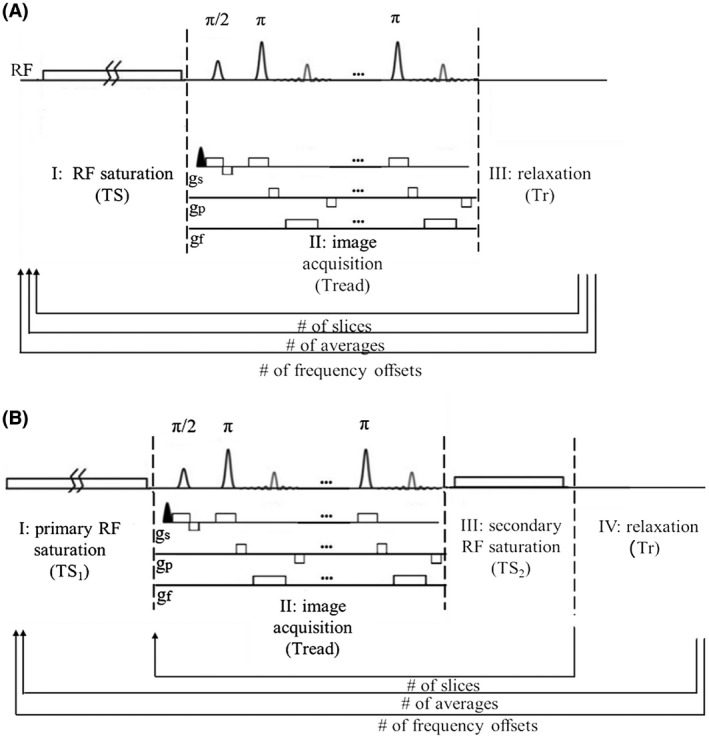
Comparison of the conventional 2D sequence and the proposed 3D multislice sequence. A, The 2D conventional sequence includes a long saturation module followed by a fast rapid acquisition with refocusing echoes (RARE) readout. The sequence is iterated for each frequency offset and for the number of slices. B, The 3D multislice sequence segments the RF irradiation in a long saturation period and a short repetitive period. The long irradiation is repeated only when the offset changes.
The total scan time for every frequency offset can be calculated as
| (1) |
and
| (2) |
for the conventional and proposed scheme, respectively, where Tr is the presaturation delay, NA/NS is the number of averages/slices, and Tread is the readout time.
The proposed fast multislice CEST‐MRI RARE‐based sequence was implemented on three different Bruker ParaVision versions (5.1, 6.0 and 360), and the implemented code is available on request to the corresponding author.
2.2. Parameter simulation/selection of optimal saturation conditions for iopamidol
Iopamidol CEST spectra were numerically simulated using a numerical Bloch‐McConnell equation solver as described in Zaiss et al. 60 , 61 The Bloch‐McConnell system consisted of six pools: water and semisolid magnetization‐transfer pool and four CEST pools of iopamidol. The water and magnetization‐transfer pool parameters were chosen as tissue‐like exchange rates for iopamidol, and chosen for a physiological condition at pH 7.1. 62 Detailed Bloch simulation parameters are given in Table 1. The RARE readout was emulated by assuming the initial magnetization before the saturation block to be small: Mz = 0.1*M0. Outgoing from a continuous block pulse (number of pulses N = 1) of duration 5 seconds, the B1 was manually optimized to yield strong CEST effects for both the 4.3‐ppm and the 5.5‐ppm resonance. With the optimal B1 fixed, the duration of the saturation (TS) was optimized. These two steps were repeated until convergence. Moreover, the influence of different T1 for each B1 and TS combinations was investigated. The T1 ranged from 1, 1.2, 1.5, 2, to 3, according to expected in vivo T1 values. 63 , 64 , 65
TABLE 1.
Tissue‐like Bloch‐McConnell simulation parameters for a iopamidol fraction of 30 mM/111 M = 1/3700 at pH = 7.1
| Water pool | MT pool | Iopamidol 1 | Iopamidol 2 | Iopamidol 3 | Iopamidol 4 | |
|---|---|---|---|---|---|---|
| Chemical shift | 0 ppm | −2.6 ppm | 5.5 ppm | 4.3 ppm | 1.8 ppm | 0.8 ppm |
| Fraction f | 1 | 0.05 | 1/3700 | 2/3700 | 1/3700 | 4/3700 |
| R1 | 0.5 Hz | 0.5 Hz | – | – | – | – |
| R2 | 28 Hz | 109 890 Hz | 66.66 Hz | 66.66 Hz | 66.66 Hz | 66.66 Hz |
| Exchange rate | 40 Hz | 1200 | 380 | 1104 | 1715 |
Abbreviation: MT, magnetization transfer.
3. METHODS
Phantom experiments using iopamidol solutions at several pHs and concentrations values were prepared to test the proposed sequence. Single‐slice and multislice acquisitions were performed in the same sample to compare different acquisition strategies. In vivo experiments in healthy and tumor‐bearing mice were performed to evaluate the simulated saturation parameters and for validating the proposed sequence for whole‐organ acquisition and pH imaging.
3.1. Phantoms
Two iopamidol‐containing phantoms were used: one (phantom #1) containing 30‐mM iopamidol solution titrated at pH 6.7, and the other (phantom #2) containing seven vials of 30‐mM iopamidol solution dissolved in phosphate‐buffered saline 1 with pH in the range of 5.5‐7.9. For both phantoms, gadolinium was added to adjust the T1 to 1.5 seconds and 1.9 seconds for phantoms #1 and #2, respectively (Supporting Information Figure S1).
3.2. Cell culture
A murine, metastatic mammary carcinoma 4T1 cell line was used for the in vivo experiments. Cells were grown in Roswell Park Memorial Institute 1640 medium containing 10% (vol/vol) fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin. Roswell Park Memorial Institute 1640 medium, fetal bovine serum, and trypsin were purchased from Lonza (Verviers, Belgium). The penicillin‐streptomycin mixture was purchased from Sigma Chemical (St. Louis, MO). Cells were incubated in 75‐cm2 flasks in a humidified 5% CO2 incubator at 37°C. At confluence, 4T1 cells were detached by adding 1 mL of trypsin–ethylene diamine tetraacetic acid solution (0.25% [wt/vol] trypsin/0.53 mM ethylene diamine tetraacetic acid).
3.3. Animal model
All animal procedures and husbandry were performed in accordance with European guidelines under directive 2010/63 and under the approval of the Italian Ministry of Health. All mice were purchased from Envigo (San Pietro al Natisone, Italy) and housed in a temperature‐controlled room with a 12‐hour light/12‐hour dark schedule and fed with autoclaved chow and water ad libitum. The in vivo imaging studies were performed on two cohorts of animals based on tumor or kidney imaging acquisition. In the first group, 8‐to‐10‐week‐old female Bagg albino mice (Charles River Laboratories Italia, Calco, Italy) were subcutaneously inoculated into both flanks with 4.0 × 105 4T1 cells, and imaging acquisition was performed when tumors reached 5‐7 mm in diameter. The second group of animals included 8‐to‐10‐week‐old male healthy Bagg albino mice for imaging of kidneys.
During the MRI experimental setup, mice were maintained under systemic anesthesia provided by the mixture of tiletamine/zolazepam 20 mg/kg (Zoletil 100; Virbac, Milan, Italy) and xylazine 5 mg/kg (Rompum; Bayer, Milan, Italy), injected intramuscularly.
3.4. Pulse sequence
3.4.1. Saturation parameter optimization
The proposed multislice sequence and optimal parameters were validated experimentally in terms of CEST contrast and pH sensitivity by testing different saturation conditions (saturation power levels and secondary saturation times or TS2). Primary pulse duration (TS1) was fixed to 3 seconds. Saturation‐pulse power levels were varied among 3.5, 3.0, and 2.5 µT, and two different TS2 of 1 and 0.5 seconds were selected for each power level. Z‐spectra were acquired on phantom #2 with RF irradiation applied at 181 frequency offsets unevenly distributed from −10 to 10 ppm relative to the water signal at 7 T and 310 K. Other parameters of the imaging protocol were TR = 11 256 ms, TE = 3.87 ms, matrix = 64 × 64, FOV = 30 × 30 mm2, slice thickness = 1.5 mm, slice gap = 0 mm, and number of slices = 8. Partial Fourier sampling was also considered, with a partial Fourier sampling Factor (PFF) of 1.6.
3.4.2. Single‐slice and multislice approach comparison
The in vitro performance of the proposed optimized scheme was validated by comparison with a conventional single‐slice acquisition sequence with the same readout. All images were acquired at 7 T on phantom #1. For the multislice sequence, the following parameters were used: B1 = 3 µT, TS1 = 3 seconds, TS2 = 1 second, TR/TE = 11 256/3.87 ms, matrix = 64 × 64, FOV = 30 × 30 mm2, number of slices = 8, slice thickness = 1.5 mm and PFF = 1.6, resulting in a readout duration of 157 ms per slice. Acquisition with the single‐slice sequence was done for comparison with the following parameters: B1 = 3 µT, TS = 5 seconds, TR = 6000 ms, TE = 3.49 ms, matrix = 64 × 64, FOV = 30 × 30 cm2, slice thickness = 1.5 mm, and PFF = 1. Eight contiguous slices were collected sequentially from bottom to top, iterating the use of the sequence along the z‐direction, to cover the entire volume of interest. For both the sequences, a multi‐offsets protocol of 181 frequencies distributed in the range of −10 to 10 ppm relative to the water resonance was applied. Acquired Z‐spectra were compared for both sequences, and the calculated CEST contrast along the slices was also evaluated. The two sequences were also compared in terms of CEST contrast‐to‐noise ratio, calculated as the ratio between the CEST contrast and CEST contrast SD.
3.4.3. Pulse sequence validation
Sequence stability along slices and accuracy to measure pH were validated in vitro on phantom #2 at 7 T and 310 K, using the previously described parameters.
3.5. Magnetic resonance imaging in vivo experiments
All images were acquired at 7 T. The parameters of B1 = 3 µT, TS1 = 3 seconds, and TS2 = 1 second were selected for the saturation module. The image matrix was 64 × 64 for a FOV of 30 × 30 mm2, with a slice thickness of 1.5 mm and eight slices. Other acquisition parameters were PFF = 1.6, TR = 11 256 ms, and TE = 3.87 ms.
For 3D in vivo CEST experiments, mice were anesthetized as previously reported, and a tail vein catheter for iopamidol injection was placed. The respiratory rate was monitored using an air pillow (SA Instruments, Stony Brook, NY). After the acquisition of scout images and a T2‐weighted anatomical reference image, Z‐spectra before and after iopamidol injection (dose: 4 g iodine/kg body weight for tumor pH imaging and 1.5 g iodine/kg body weight for kidney pH imaging, respectively) were acquired and analyzed to calculate the 3D pH maps. For each CEST acquisition, 46 frequency offsets were acquired by varying the center frequency of the CEST RF pulse from −10 to 10 ppm with respect to the water signal.
3.6. Data analysis
All data were processed using custom‐written scripts in MATLAB R2015b (MathWorks, Natick, MA).
For CEST analysis, images were elaborated as described in Terreno et al. 66 Briefly, a smoothed splines algorithm was used to interpolate the Z‐spectrum. The corresponding B0 field shifting was calculated as the distance of the minimum of the interpolated spectrum from 0 ppm. To correct the B0 inhomogeneity effect, the minimum position was exploited to shift the offset axis, and the new corrected spectrum was calculated by cubic smoothing spline interpolation using the shifted offsets vector as input. The CEST contrast was quantified at a specific offset, using the asymmetry analysis:
| (3) |
where S ±Δ ω is the water signal intensity in the presence of a saturation pulse at offset ±Δω (ie, Δω = 4.2/5.5 ppm for iopamidol). Ratiometric values were calculated by dividing the different contrast values obtained at the two different offsets, according to the following equation:
| (4) |
Voxel‐wise pH maps were obtained by extrapolating the pH values from the previously obtained ratiometric curves. 27 For the in vivo images, difference CEST contrast maps (ΔST) were calculated at 4.2 ppm and 5.5 ppm as the difference between the saturation‐transfer contrast after and before iopamidol injection on a voxel‐wise basis, to retain the CEST signals arising only from the contrast agent and to eliminate CEST endogenous contributions. A threshold value of 1% was set, based on the scanner signal fluctuations, to discriminate between enhancing and nonenhancing pixels following iopamidol injection. From the ΔST maps at the two frequencies, the corresponding pH maps were then calculated.
4. RESULTS
4.1. Segmented multislice sequence
The sequence schemes for the single‐slice and the multislice acquisitions are depicted in Figure 1. The conventional single‐slice sequence based on a centric‐encoded RARE readout includes three periods: relaxation recovery, RF irradiation, and single‐shot fast spin‐echo acquisition. Even when using fast single‐shot image acquisition schemes, multislice images can only be obtained with very long acquisition times due to the repeated saturation periods for each offset and slice. In contrast, the proposed sequence exploits an identical fast single‐shot RARE readout, but the saturation module is split into two parts with different durations: the first based on a longer saturation period and the second on a shorter one, repeated before each slice acquisition. Because the slices are acquired just after the second short saturation pulse rather than following each long first saturation period, the total scan time for the whole organs is dramatically reduced.
4.2. Optimization and validation of saturation parameters
In Figure 2, tissue‐like Bloch‐McConnell simulations for iopamidol are shown. The inclusion of direct water saturation and semisolid magnetization‐transfer background leads to a relatively low optimal saturation power of 3 µT to yield strong CEST effects for both resonances (Figure 2A,E,I). Moreover, at this power level, the CEST effects for both resonances already decrease for saturation durations longer than 3 seconds, and for a saturation period longer than 0.5 seconds, is still greater or equal to 75% of the effect at 10 seconds of saturation (Figure 2B,F,J). Therefore, by using optimized B1, only 3 seconds of saturation duration and a short recovery time yield the same CEST contrast as longer schemes and, considering that the short secondary pulse needs to recover saturation losses due to the acquisition time (readout time ~160 ms), a short saturation (TS2 of 0.5 second) can be enough to ensure a good CEST contrast in the whole volume. In Figure 2D,H,L, the CEST effect at different T1 (R1) values is simulated. Interestingly, for T1 values in the range observed in vivo (1‐3 seconds), the CEST contrast is almost not affected by the exploited saturation times.
FIGURE 2.
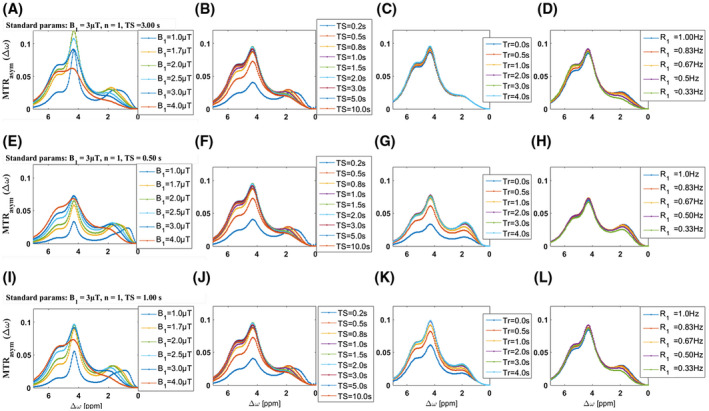
Numerical optimization of saturation parameters. Optimization of saturation power B1 (A,E,I), saturation pulse duration (TS) (B,F,J) and recovery time (Tr) before saturation (C,G,K), and magnetization transfer rate asymmetry (MTRasym) simulation at different R1 (D,H,L). Standard parameters if unchanged were 3 µT, TS = 3 seconds, and Tr = 5 seconds (A‐D); B1 = 3 µT, TS = 0.5 seconds, and Tr = 5 seconds (E‐H); and B1 = 3 µT, TS = 0.5 seconds, and Tr = 5 seconds (I‐L).
These simulated results were verified in vivo by single‐slice acquisitions. Breast tumor–bearing mice showed similar contrast enhancements following iopamidol injection for both the amide groups at 4.2 ppm and 5.5 ppm independently of the shorter (3‐second) or longer (5‐second) saturation schemes (Figure 3A,B). Moreover, a similar extravasation fraction was observed when applying the short (3‐second) saturation period (Figure 3C). Representative single‐slice CEST contrast maps superimposed to anatomical images are shown for iopamidol at 4.2 ppm (Figure 3D) and 5.5 ppm (Figure 3E) after 5 seconds or 3 seconds of saturation duration (Figure 3F,G).
FIGURE 3.
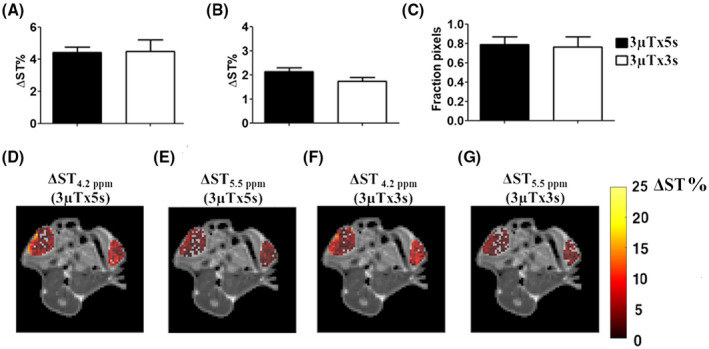
In vivo validation of simulated saturation schemes. Average ΔST% contrast measured in tumor regions at 4.2 ppm (A) and at 5.5 ppm (B) and corresponding fraction pixels (C) following iopamidol injection for the two investigated irradiation conditions (3 µT × 5 seconds and 3 µT × 3 seconds, respectively) show similar contrast efficiency. Representative single‐slice ΔST contrast‐enhanced maps superimposed to T2‐weighteed anatomical images for the same tumor‐bearing mouse obtained following saturation with the same power (3 µT) but different durations: 5 seconds at 4.2 ppm and 5.5 ppm (D,E), and 3 seconds at 4.2 ppm and 5.5 ppm (F,G).
4.3. In vitro optimization of pH sensitivity
Next, we investigated the effect of different saturation conditions on the measured pH responsiveness. Figure 4 shows the CEST contrast obtained with the proposed multislice sequence at different saturation powers B1 and secondary saturation pulse durations TS2 as a function of the pH. As shown in Figure 4A,B (solid lines), the CEST contrast increases with pH, peaking at pH = 6.7 for the resonance at 5.5 ppm and at pH = 7.4 for the resonance at 4.2 ppm. For B1 = 3.5 µT (Figure 4A, red solid line), the maximum value was reached at the highest pH value. As expected, experiments in which B1 was incremented showed an increase in the maximum contrast for both frequency offsets. In particular, the CEST contrasts calculated at 4.2 ppm, pH = 7, and secondary saturation duration of 1 second were 17.7%, 22.2%, and 26.3% for B1 = 2.5 µT, 3 µT, and 3.5 µT, respectively, whereas the CEST contrast measured at 5.5 ppm, pH = 6.7, and at the same saturation condition was 8.0%, 10.2%, and 13.5%, respectively. Similar behavior was observed for TS2 = 0.5 seconds (dashed lines), but contrast values were generally lower than the ones obtained at the same power and with a longer secondary saturation period. Figure 4C (solid lines) illustrates the CEST ratiometric values as a function of pH at different B1 and TS2 values. The ratiometric curve linearly increased with the pH values but showed higher pH sensitivity (ie, increased steepness) for lower powers. When reducing TS2, the ratiometric curves were almost similar (Figure 4C, dashed lines). Therefore, the optimal saturation conditions were determined as the best tradeoff between the maximum detectable CEST contrast at the two frequencies and the highest pH sensitivity, corresponding to the following conditions: saturation power = 3 µT, TS1 = 3 seconds, and TS2 = 1 second.
FIGURE 4.
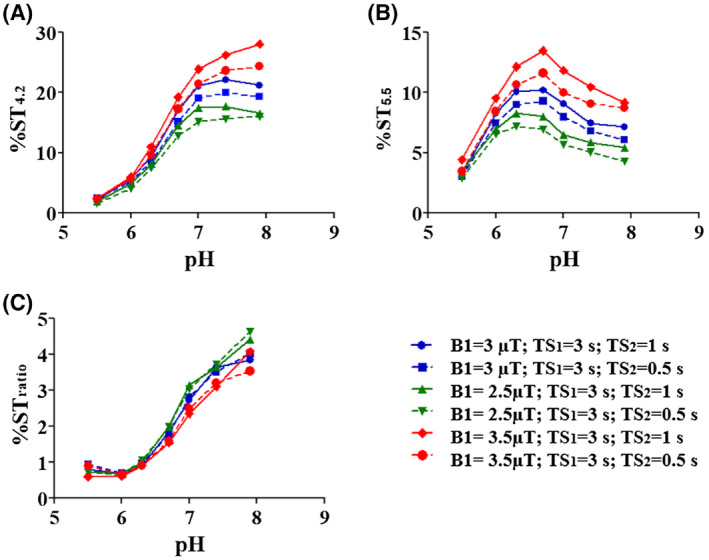
Saturation parameters optimization. The CEST contrast calculated at 4.2 ppm (A) and 5.5 ppm (B) and CEST ratio (C) as a function of pH for different saturation conditions.
4.4. Single‐slice and multislice sequence comparison
Figure 5 compares the single‐slice and the proposed multislice sequences. In particular, Figure 5A shows the Z‐spectra of phantom #1 (pH = 6.7), acquired by iterating the conventional single‐slice RARE sequence (TS = 5 seconds) over the slices in a contiguous way. In comparison, Figure 5B shows the same Z‐spectra collected with the optimized multislice sequence (TS1 = 3 seconds; TS2 = 1 second). In both the acquisition schemes the Z‐spectra are fully overlapped for all eight slices. The relative differences between each slice and the mean volumetric Z‐spectrum showed percentage variations in the range ±0.9% for the single‐slice sequence (Figure 5C) and ±1.3% for the multislice one (Figure 5D), with comparable changes versus slices. Moreover, the two sequences were compared for image quality and quantitatively in terms of CEST contrast. Figure 5E,F shows the acquired images at 4.2 ppm for the third slice: for both approaches, the images appear well defined, with no evident artifacts or geometrical distortions, exploiting one of the RARE readout advantages. The ST% maps for slice 3 are shown in Figure 5G,H. The average ST% at 4.2 ppm and 5.5 ppm were 8.25 ± 0.4% and 10.4% ± 0.4% for the single‐slice approach, whereas they were 8.4% ± 0.7% and 10.7% ± 0.6% for the multislice acquisition scheme. Additional images for all slices for the two sequences are shown in the Supporting Information Figure S2, and the corresponding CEST contrast maps at 4.2 ppm and at 5.5 ppm are shown in Supporting Information Figure S3. These findings highlight that the measured CEST contrasts, even when using shorter saturation periods, compare with those obtained with the conventional single‐slice approach. The CEST contrast‐to‐noise ratio was evaluated along the slices for both methods (Figure 5I). The contrast‐to‐noise ratio mean values were 16.9 ± 0.9 and 12.0 ± 1.4 for the single‐slice and multislice approaches, respectively. The calculated contrast‐to‐noise ratio was lower for the multislice approach when compared with the single‐slice approach, but still high and homogeneous along the slices. Moreover, by using the proposed sequence, the total acquisition time for a single Z‐spectrum sampled with 46 offsets in eight slices covering a volume of 30 × 30 × 12 mm3 with a spatial resolution of 0.47 × 0.47 × 1.5 mm3 was 9 minutes and 29 seconds for the multislice scheme compared with 36 minutes and 48 seconds for the conventional approach (repeated single‐slice acquisition).
FIGURE 5.

Comparison of Z‐spectra signal intensities, percentage variations with respect to the average Z‐spectra along the slices, acquired T2‐weighted image at 4.2 ppm and calculated CEST contras at 4.2 ppm and 5.5 ppm obtained with the conventional 2D single‐slice sequence (A,C,E,G) and the proposed 3D multislice approach (B,D,F,H). Z‐spectra nearly overlay for all of the slices and provided similar CEST contrast for both methods. I, The calculated contrast‐to‐noise ratio (CNR) along the slices for the two investigated acquisition approaches.
4.5. In vitro sequence validation
Next, we evaluated the homogeneity along the slices for the proposed multislice scheme in terms of both CEST contrast and pH. Figure 6 shows the results of the validation experiments on the pH‐varying iopamidol phantom #2. The 2D multislice CEST contrast maps at 4.2 ppm and 5.5 ppm are shown in Figure 6A,C, and the corresponding average calculated saturation‐transfer values for each vial are shown in Figure 6B,D. The measured saturation‐transfer values for both the 4.2‐ppm and 5.5‐ppm pools were found to be constant along all of the slices, with a clear dependence on pH but not on the position along the main magnetic axis. Linear regression analysis showed that the slope was not significantly different from zero for all vials; hence, the measured CEST contrast is not affected by the slice position. From the measured CEST contrast at the two frequencies, we calculated the corresponding pH values across all of the slices (Figure 7). The calculated pH maps are very homogenous along the slice direction within the same vial (Figure 7A). The average calculated pH values for each vial (in the pH range 6.0‐7.4) showed no differences across all slices (Figure 7B). As for the contrast, the pH linear regression analysis also showed that the slope was not significantly different from zero; therefore, the proposed multislice sequence allows one to obtain accurate pH measurements for all slices. Moreover, very high correlations were observed between the measured (pH meter) and CEST‐calculated pH values (Figure 7C) for all slices. The calculated coefficients of determination are summarized for each slice in Supporting Information Table S1 and they ranged from 0.94 to 0.99. To further characterize the pH measure accuracy along the z‐direction, mean pH values along the slices were calculated and compared with the experimental ones (Figure 7D). Mean calculated pH values were very similar to the experimental ones with low SDs, confirming the accuracy along the third dimension. Moreover, Pearson’s correlation coefficient and the coefficient of determination between calculated and experimental pH values were very high (0.995 and 0.991, respectively).
FIGURE 6.

In vitro validation. Two‐dimensional multislice contrast map of the pH‐varying iopamidol phantom #2 at 4.2 ppm (A) and 5.5 ppm (C). Mean CEST contrast for each vial as a function of the slice position (points) and corresponding regression lines (dashed lines) at 4.2 ppm (B) and 5.5 ppm (D).
FIGURE 7.

Assessment of sequence accuracy in measuring pH. A, Two‐dimensional multislice pH map of the pH‐varying iopamidol phantom #2. B, Mean compartmental pH for each vial as a function of the slice position (points) and corresponding regression lines (dashed lines). C, Correlation between calculated pH and real pH for each slice. D, Correlation between average calculated pH and real pH values.
4.6. In vivo sequence validation
The capability of the proposed multislice sequence to measure pH in whole organs was evaluated in vivo in two different cases: tumors and kidneys. The sequence allowed us to acquire eight slices in less than 10 minutes by sampling 46 frequency offsets. Moreover, considering the low mean B0 shift (see Supporting Information Figure S4), the acquisition time could be potentially further decreased by reducing the number and the range of the sampled frequencies. Calculated pH parametric maps superimposed to anatomical T2w images are shown for a representative tumor‐bearing mouse (Figure 8) and for the kidneys for a healthy mouse (Figure 9). The multislice acquisition allows the 3D rendering of the corresponding pH maps, which helps in the visualization of the pH distribution within the whole organ (Figures 8B and 9B). In addition, the information conveyed by the 3D data sets can be exploited for assessing pH gradients inside the organs. Of note, pH values in the whole tumors were observed to be less acidic in the rim regions and more acidic in the core region (Figure 8C). For the kidneys, we observed a neutral to acidic pH gradient when moving from the cortex to the medulla and the calyx (Figure 9C).
FIGURE 8.
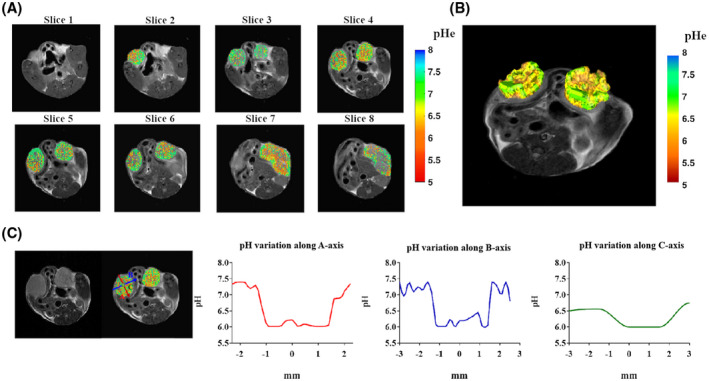
A, Two‐dimensional multislice tumor extracellular pH (pHe) map for a breast‐tumor murine model. B, Three‐dimensional pH map rendering. C, Calculated pH gradients along the three main axes inside the left tumor region showing T2‐weighted image, overimposed pH map, and pH gradients along the A‐axis (red), B‐axis (blue), and C‐axis (green).
FIGURE 9.

A, Two‐dimensional multislice pH map of the kidneys of a healthy mouse. B, Three‐dimensional pH map rendering. C, Calculated pH gradients along the three main axes inside the right kidney showing T2‐weighted image, overimposed pH map, and pH gradients along A‐axis (red), B‐axis (blue), and C‐axis (green).
5. DISCUSSION
In this work we developed a segmented multislice CEST sequence based on a fast single‐shot RARE readout for in vivo imaging pH in whole organs. The proposed sequence provided a homogeneous CEST contrast along the slices through the repetition of a short saturation pulse before each slice acquisition, therefore increasing the CEST‐MRI efficiency without significantly prolonging the acquisition time. Moreover, the single‐shot RARE readout and the partial Fourier sampling speed up the image acquisition without introducing visible artifacts. 67 As a result, our protocol obtained CEST Z‐spectra sampled at 46 frequencies in eight slices within 9 minutes and 29 seconds, which equates to about 12 seconds/volume/offset or to 1.5 seconds/offset/slice.
Because of the importance of whole‐organ volume coverage, several volumetric sequences have been developed for both clinical and preclinical scanners. At the clinical level, most of the sequences use 2D/3D approaches and exploit parallel imaging to obtain very rapid acquisitions with an acquisition time per offset that varies from 4 seconds to 20 seconds, according to the saturation duration and the selected readout. 41 , 43 , 44 , 45 , 46 , 49 , 50 , 68 Echo planar imaging and gradient and spin echo–based sequences are very fast and efficient, but they can be affected by physiological noise and geometric distortion. 43 , 46 , 68 Gradient‐echo readout shows high signal stability but may suffer from some image artifacts. 41 , 49 A RARE readout overcomes these issues but is usually slower and can have specific absorption rate limitations. 44 , 50 At the preclinical level, instead, few multislice schemes have been developed but not yet integrated into any of the commercial MRI scanners for animal experimentations. Sun et al 55 proposed an EPI‐based CEST sequence that allows covering a FOV of 25 × 25 × 10 mm3 with a spatial resolution of 0.5 × 0.5 × 2 mm3 in 6.5 seconds/offset (~5‐minute total acquisition duration) at the expense of an additional T1 map acquisition to compensate for T1‐related contrast losses. A GRE readout scheme has been proposed by Dixon et al by repeating the saturation period before the acquisition of each slice, but the repetition generates a cumulative effect on the contrast that depends on the final slice number. 56 Sun et al 57 demonstrated that an uneven segmentation of the saturation periods coupled with a GRE readout provides a fast and efficient CEST acquisition scheme of approximately 3.5 seconds/offset (FOV = 25 × 25 × 10 mm3, spatial resolution = 0.37 × 0.37 × 2 mm3, and 48 frequency offsets in about 4 minutes). A CEST‐FISP POMP approach was developed with high‐volume coverage in a short time, thanks to the simultaneous acquisition of multiple slices. 37 This sequence allows covering a volume of 60 × 60 × 10 mm3 with an isotropic spatial resolution of 0.5 mm3 in 7 minutes and 28 seconds, which equates to 0.9 seconds/slice/frequency. Compared with the previously developed multislice sequences, our approach shows several potential advantages, including (1) fewer susceptibility artifacts and greater SNR than EPI‐based and GRE‐based schemes; (2) no requirement of a T1 map for proper relaxation correction, in contrast to the EPI‐based sequence; (3) no requirement of a dedicated multichannel hardware as for parallel imaging, which is not usually available in preclinical scanners; and (4) reduced B0 inhomogeneities and higher pH accuracy (0.99 against 0.83 for calculated vs experimental pH values) in comparison to the POMP scheme.
In this work, the optimization of our proposed fast multislice scheme sequence aimed at improving both CEST contrast efficiency and pH accuracy. We demonstrated by simulation and in vivo studies that optimal saturation conditions were achieved for iopamidol by exploiting a continuous‐wave saturation pulse of 3 µT with a duration as short as 3 seconds. Moreover, we observed that the highest pH responsiveness can be achieved with a tradeoff between CEST contrast and scan duration, resulting in a segmented saturation period of 1 second per each slice (TS2), which is sufficient to maintain the contrast without signal losses along the slices. We found that both Z‐spectra and CEST contrasts obtained with the multislice approach were homogeneous along slices and comparable to the values obtained using the single‐slice approach. These results demonstrate that this segmented multislice approach offers image quality comparable to that achievable with the single‐slice approach, with a significant time savings. We validated the accuracy of measuring pH along all three dimensions in pH‐titrated phantoms. In vitro results showed a good pH homogeneity along the z‐dimension and a significant and high correlation between calculated and real pH, hence reflecting high precision for measuring pH.
The availability of whole‐volume coverage sequences allows one to obtain more accurate pH‐related information associated with cancer metabolism or renal functionality. It is well documented that tumor acidity has profound consequences on tumor progression and that tumor pH heterogeneity further promotes aggressiveness and invasiveness of cancer cells. 69 , 70 , 71 The calculated tumor pH maps showed this heterogeneity in the whole tumor, with a marked gradient (from less to more acidic) between the rim and the core areas, likely reflecting the reduced perfusion in the inner tumor regions, hence higher glycolytic activity. 23 Three‐dimensional pH maps in kidneys also showed a pH gradient, with neutral pH values in the cortex and more acidic pH values in the medulla and the calyx regions, in agreement with renal physiology. A similar pH distribution was previously reported, suggesting that our approach can be used to further assess whole‐kidney pH. 27 , 72 , 73
6. CONCLUSIONS
Our proposed sequence based on a segmented irradiation scheme coupled with a single‐shot centric RARE readout provides fast acquisition and good contrast without geometrical distortions. The optimization of the saturation scheme resulted in good Z‐spectra homogeneity along the slices, comparable to the conventional single‐slice approach, but with a significant reduction in the overall acquisition time. Very high accuracy in measuring pH was observed in a phantom with pH values in the physiological range along the three dimensions. These results show that the proposed multislice CEST sequence can offer a new tool to investigate the 3D pH heterogeneity in whole organs, both in pathological and physiological conditions.
Supporting information
FIGURE S1 T1 maps for phantom #1 (A) and phantom #2 (B)
FIGURE S2 The CEST images at 4.2, 5.5, and 10 ppm for single‐slice (A,C,E) and multislice acquisition (B,D,F)
FIGURE S3 Saturation‐transfer CEST maps at 4.2 ppm and 5.5 ppm for the single‐slice (A,C) and the multislice (B,D) approach
FIGURE S4 B0 shift maps (A) and mean B0 shift region of interest–averaged values for the tumor‐bearing mouse shown in Figure 8A,B and for the kidneys of the healthy mouse shown in Figure 9C,D
TABLE S1 Coefficient of determination for the measured and calculated pH values for phantom #2
ACKNOWLEDGMENTS
This work was supported by grants from the Associazione Italiana Ricerca Cancro (AIRC MFAG 2017 ‐ ID. 20153 project – P.I. Longo Dario Livio), from the Compagnia San Paolo project (Regione Piemonte; Grant No. CSTO165925), and from the European Union’s Horizon 2020 research and innovation program (Grant No. 667510). Lorena Consolino was supported by the Fondazione Umberto Veronesi fellowship program. The Italian Ministry for Education and Research is gratefully acknowledged for its yearly FOE funding to the Euro‐BioImaging Multi‐Modal Molecular Imaging Italian Node.
Villano D, Romdhane F, Irrera P, et al. A fast multislice sequence for 3D MRI‐CEST pH imaging. Magn Reson Med. 2020;85:1335–1349. 10.1002/mrm.28516
Funding information
Associazione Italiana Ricerca Cancro (AIRC MFAG 2017 ‐ ID. 20153 project – P.I. Longo Dario Livio); Compagnia San Paolo project (Regione Piemonte; Grant No. CSTO165925); and the European Union’s Horizon 2020 research and innovation program (Grant No. 667510)
DATA AVAILABILITY STATEMENT
The developed MRI‐CEST multislice sequence is freely available at https://www.cim.unito.it/website/research/research_MRI_seq.php.
REFERENCES
- 1. van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): What is in a name and what isn't? Magn Reson Med. 2011;65:927‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu G, Song X, Chan KW, McMahon MT. Nuts and bolts of chemical exchange saturation transfer MRI. NMR Biomed. 2013;26:810‐828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terreno E, Castelli DD, Aime S. Encoding the frequency dependence in MRI contrast media: The emerging class of CEST agents. Contrast Media Mol Imaging. 2010;5:78‐98. [DOI] [PubMed] [Google Scholar]
- 4. Zhang S, Malloy CR, Sherry AD. MRI thermometry based on PARACEST agents. J Am Chem Soc. 2005;127:17572‐17573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McVicar N, Li AX, Suchy M, Hudson RH, Menon RS, Bartha R. Simultaneous in vivo pH and temperature mapping using a PARACEST‐MRI contrast agent. Magn Reson Med. 2013;70:1016‐1025. [DOI] [PubMed] [Google Scholar]
- 6. Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J Magn Reson Imaging. 2018;47:11‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He Y‐L, Li Y, Lin C‐Y, et al. Three‐dimensional turbo‐spin‐echo amide proton transfer‐weighted MRI for cervical cancer: A preliminary study. J Magn Reson Imaging. 2019;50:1318‐1325. [DOI] [PubMed] [Google Scholar]
- 8. Jones KM, Randtke EA, Yoshimaru ES, et al. Clinical translation of tumor acidosis measurements with AcidoCEST MRI. Mol Imaging Biol. 2017;19:617‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meissner J‐E, Korzowski A, Regnery S, et al. Early response assessment of glioma patients to definitive chemoradiotherapy using chemical exchange saturation transfer imaging at 7 T. J Magn Reson Imaging. 2019;50:1268‐1277. [DOI] [PubMed] [Google Scholar]
- 10. Kogan F, Hariharan H, Reddy R. Chemical exchange saturation transfer (CEST) imaging: Description of technique and potential clinical applications. Curr Radiol Rep. 2013;1:102‐114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zaric O, Farr A, Poblador Rodriguez E, et al. 7T CEST MRI: A potential imaging tool for the assessment of tumor grade and cell proliferation in breast cancer. Magn Reson Imaging. 2019;59:77‐87. [DOI] [PubMed] [Google Scholar]
- 12. Jones CK, Schlosser MJ, van Zijl PC, Pomper MG, Golay X, Zhou J. Amide proton transfer imaging of human brain tumors at 3T. Magn Reson Med. 2006;56:585‐592. [DOI] [PubMed] [Google Scholar]
- 13. Togao O, Hiwatashi A, Keupp J, et al. Amide proton transfer imaging of diffuse gliomas: Effect of saturation pulse length in parallel transmission‐based technique. PLoS One. 2016;11:e0155925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wen Z, Hu S, Huang F, et al. MR imaging of high‐grade brain tumors using endogenous protein and peptide‐based contrast. Neuroimage. 2010;51:616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61:1441‐1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee D‐H, Lee D‐W, Kwon J‐I, et al. In vivo mapping and quantification of creatine using chemical exchange saturation transfer imaging in rat models of epileptic seizure. Mol Imaging Biol. 2019;21:232‐239. [DOI] [PubMed] [Google Scholar]
- 18. Kogan F, Haris M, Singh A, et al. Method for high‐resolution imaging of creatine in vivo using chemical exchange saturation transfer. Magn Reson Med. 2014;71:164‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange‐dependent saturation transfer (gagCEST). Proc Natl Acad Sci USA. 2008;105:2266‐2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juras V, Winhofer Y, Szomolanyi P, et al. Multiparametric MR imaging depicts glycosaminoglycan change in the achilles tendon during ciprofloxacin administration in healthy men: Initial observation. Radiology. 2015;275:763‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Müller‐Lutz A, Khalil N, Schmitt B, et al. Pilot study of Iopamidol‐based quantitative pH imaging on a clinical 3T MR scanner. MAGMA. 2014;27:477‐485. [DOI] [PubMed] [Google Scholar]
- 22. Consolino L, Anemone A, Capozza M, et al. Non‐invasive investigation of tumor metabolism and acidosis by MRI‐CEST imaging. Front Oncol. 2020;10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Longo DL, Bartoli A, Consolino L, et al. In vivo imaging of tumor metabolism and acidosis by combining PET and MRI‐CEST pH imaging. Cancer Res. 2016;76:6463‐6470. [DOI] [PubMed] [Google Scholar]
- 24. Anemone A, Consolino L, Conti L, et al. In vivo evaluation of tumour acidosis for assessing the early metabolic response and onset of resistance to dichloroacetate by using magnetic resonance pH imaging. Int J Oncol. 2017;51:498‐506. [DOI] [PubMed] [Google Scholar]
- 25. Chen LQ, Randtke EA, Jones KM, Moon BF, Howison CM, Pagel MD. Evaluations of tumor acidosis within in vivo tumor models using parametric maps generated with acido CEST MRI. Mol Imaging Biol. 2015;17:488‐496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anemone A, Consolino L, Arena F, Capozza M, Longo DL. Imaging tumor acidosis: A survey of the available techniques for mapping in vivo tumor pH. Cancer Metastasis Rev. 2019;38:25‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Longo DL, Dastrù W, Digilio G, et al. Iopamidol as a responsive MRI‐chemical exchange saturation transfer contrast agent for pH mapping of kidneys: In vivo studies in mice at 7 T. Magn Reson Med. 2011;65:202‐211. [DOI] [PubMed] [Google Scholar]
- 28. Longo DL, Michelotti F, Consolino L, et al. In vitro and in vivo assessment of nonionic iodinated radiographic molecules as chemical exchange saturation transfer magnetic resonance imaging tumor perfusion agents. Invest Radiol. 2016;51:155‐162. [DOI] [PubMed] [Google Scholar]
- 29. Anemone A, Consolino L, Longo DL. MRI‐CEST assessment of tumour perfusion using X‐ray iodinated agents: Comparison with a conventional Gd‐based agent. Eur Radiol. 2017;27:2170‐2179. [DOI] [PubMed] [Google Scholar]
- 30. High RA, Randtke EA, Jones KM, et al. Extracellular acidosis differentiates pancreatitis and pancreatic cancer in mouse models using acidoCEST MRI. Neoplasia. 2019;21:1085‐1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longo DL, Busato A, Lanzardo S, Antico F, Aime S. Imaging the pH evolution of an acute kidney injury model by means of iopamidol, a MRI‐CEST pH‐responsive contrast agent. Magn Reson Med. 2013;70:859‐864. [DOI] [PubMed] [Google Scholar]
- 32. Longo DL, Cutrin JC, Michelotti F, Irrera P, Aime S. Noninvasive evaluation of renal pH homeostasis after ischemia reperfusion injury by CEST‐MRI. NMR Biomed. 2017;30:e3720. [DOI] [PubMed] [Google Scholar]
- 33. Irrera P, Consolino L, Cutrin JC, Zollner FG, Longo DL. Dual assessment of kidney perfusion and pH by exploiting a dynamic CEST‐MRI approach in an acute kidney ischemia‐reperfusion injury murine model. NMR Biomed. 2020;33:e4287. [DOI] [PubMed] [Google Scholar]
- 34. Pavuluri K, Manoli I, Pass A, et al. Noninvasive monitoring of chronic kidney disease using pH and perfusion imaging. Science Advances. 2019;5:eaaw8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Y, Zhou IY, Igarashi T, Longo DL, Aime S, Sun PZ. A generalized ratiometric chemical exchange saturation transfer (CEST) MRI approach for mapping renal pH using iopamidol. Magn Reson Med. 2018;79:1553‐1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. High RA, Ji Y, Ma Y‐J, et al. In vivo assessment of extracellular pH of joint tissues using acidoCEST‐UTE MRI. Quant Imaging Med Surg. 2019;9:1664‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Randtke EA, Granados JC, Howison CM, Pagel MD, Cárdenas‐Rodríguez J. Multislice CEST MRI improves the spatial assessment of tumor pH. Magn Reson Med. 2017;78:97‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J, Wu Y, Guo Y, Zheng H, Sun PZ. A review of optimization and quantification techniques for chemical exchange saturation transfer MRI toward sensitive in vivo imaging. Contrast Media Mol Imaging. 2015;10:163‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kogan F, Hargreaves BA, Gold GE. Volumetric multislice gagCEST imaging of articular cartilage: Optimization and comparison with T1rho. Magn Reson Med. 2017;77:1134‐1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dula AN, Asche EM, Landman BA, et al. Development of chemical exchange saturation transfer at 7 T. Magn Reson Med. 2011;66:831‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zaiss M, Ehses P, Snapshot‐CEST SK. Optimizing spiral‐centric‐reordered gradient echo acquisition for fast and robust 3D CEST MRI at 9.4 T. NMR Biomed. 2018;31:e3879. [DOI] [PubMed] [Google Scholar]
- 42. Heo H‐Y, Xu X, Jiang S, et al. Prospective acceleration of parallel RF transmission‐based 3D chemical exchange saturation transfer imaging with compressed sensing. Magn Reson Med. 2019;82:1812‐1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones CK, Polders D, Hua J, et al. In vivo three‐dimensional whole‐brain pulsed steady‐state chemical exchange saturation transfer at 7 T. Magn Reson Med. 2012;67:1579‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mugler JP. Optimized three‐dimensional fast‐spin‐echo MRI. J Magn Reson Imaging. 2014;39:745‐767. [DOI] [PubMed] [Google Scholar]
- 45. Zhu H, Jones CK, van Zijl PC, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med. 2010;64:638‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Akbey S, Ehses P, Stirnberg R, Zaiss M, Stöcker T. Whole‐brain snapshot CEST imaging at 7 T using 3D‐EPI. Magn Reson Med. 2019;82:1741‐1752. [DOI] [PubMed] [Google Scholar]
- 47. Krishnamoorthy G, Nanga RPR, Bagga P, Hariharan H, Reddy R. High quality three‐dimensional gagCEST imaging of in vivo human knee cartilage at 7 Tesla. Magn Reson Med. 2017;77:1866‐1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitt B, Zbýn S, Stelzeneder D, et al. Cartilage quality assessment by using glycosaminoglycan chemical exchange saturation transfer and (23)Na MR imaging at 7 T. Radiology. 2011;260:257‐264. [DOI] [PubMed] [Google Scholar]
- 49. Deshmane A, Zaiss M, Lindig T, et al. 3D gradient echo snapshot CEST MRI with low power saturation for human studies at 3T. Magn Reson Med. 2019;81:2412‐2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao X, Wen Z, Zhang GE, et al. Three‐dimensional turbo‐spin‐echo amide proton transfer MR imaging at 3‐Tesla and its application to high‐grade human brain tumors. Mol Imaging Biol. 2013;15:114‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y, Heo HY, Jiang S, Bottomley PA, Zhou J. Fast, reliable 3D Amide proton transfer imaging of brain tumors at 3T with variably‐accelerated sensitivity encoding (vSENSE). In: Proceedings of the International Society for Magnetic Resonance in Medicine (Vol. 25), Honolulu, Hawaii, 2017. p. 1971. [PMC free article] [PubMed] [Google Scholar]
- 52. Khlebnikov V, Geades N, Klomp DWJ, Hoogduin H, Gowland P, Mougin O. Comparison of pulsed three‐dimensional CEST acquisition schemes at 7 Tesla: Steady state versus pseudosteady state. Magn Reson Med. 2017;77:2280‐2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Varma G, Lenkinski RE, Vinogradov E. Keyhole chemical exchange saturation transfer. Magn Reson Med. 2012;68:1228‐1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cohen O, Huang S, McMahon MT, Rosen MS, Farrar CT. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn Reson Med. 2018;80:2449‐2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun PZ, Murata Y, Lu J, Wang X, Lo EH, Sorensen AG. Relaxation‐compensated fast multislice amide proton transfer (APT) imaging of acute ischemic stroke. Magn Reson Med. 2008;59:1175‐1182. [DOI] [PubMed] [Google Scholar]
- 56. Dixon WT, Hancu I, Ratnakar SJ, Sherry AD, Lenkinski RE, Alsop DC. A multislice gradient echo pulse sequence for CEST imaging. Magn Reson Med. 2010;63:253‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun PZ, Cheung JS, Wang E, Benner T, Sorensen AG. Fast multislice pH‐weighted chemical exchange saturation transfer (CEST) MRI with Unevenly segmented RF irradiation. Magn Reson Med. 2011;65:588‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Glover GH. Phase‐offset multiplanar (POMP) volume imaging: A new technique. J Magn Reson Imaging. 1991;1:457‐461. [DOI] [PubMed] [Google Scholar]
- 59. Shah T, Lu L, Dell KM, Pagel MD, Griswold MA, Flask CA. CEST‐FISP: A novel technique for rapid chemical exchange saturation transfer MRI at 7 T. Magn Reson Med. 2011;65:432‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zaiss M, Angelovski G, Demetriou E, McMahon MT, Golay X, Scheffler K. QUESP and QUEST revisited—Fast and accurate quantitative CEST experiments. Magn Reson Med. 2018;79:1708‐1721. [DOI] [PubMed] [Google Scholar]
- 61. Zaiss M, Anemone A, Goerke S, et al. Quantification of hydroxyl exchange of D‐glucose at physiological conditions for optimization of glucoCEST MRI at 3, 7 and 9.4 Tesla. NMR Biomed. 2019;32:e4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun PZ, Longo DL, Hu W, Xiao G, Wu R. Quantification of iopamidol multi‐site chemical exchange properties for ratiometric chemical exchange saturation transfer (CEST) imaging of pH. Phys Med Biol. 2014;59:4493‐4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hueper K, Peperhove M, Rong S, et al. T1‐mapping for assessment of ischemia‐induced acute kidney injury and prediction of chronic kidney disease in mice. Eur Radiol. 2014;24:2252‐2260. [DOI] [PubMed] [Google Scholar]
- 64. Herrmann K, Johansen M, Craig S, et al. Molecular imaging of tumors using a quantitative T1 mapping technique via magnetic resonance imaging. Diagnostics (Basel). 2015;5:318‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McSheehy PM, Weidensteiner C, Cannet C, et al. Quantified tumor T1 is a generic early‐response imaging biomarker for chemotherapy reflecting cell viability. Clin Cancer Res. 2010;16:212‐225. [DOI] [PubMed] [Google Scholar]
- 66. Terreno E, Stancanello J, Longo D, et al. Methods for an improved detection of the MRI‐CEST effect. Contrast Media Mol Imaging. 2009;4:237‐247. [DOI] [PubMed] [Google Scholar]
- 67. McGibney G, Smith MR, Nichols ST, Crawley A. Quantitative evaluation of several partial Fourier reconstruction algorithms used in MRI. Magn Reson Med. 1993;30:51‐59. [DOI] [PubMed] [Google Scholar]
- 68. Zhou J, Zhu HE, Lim M, et al. Three‐dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38:1119‐1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Robertson‐Tessi M, Gillies RJ, Gatenby RA, Anderson AR. Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res. 2015;75:1567‐1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peppicelli S, Bianchini F, Calorini L. Extracellular acidity, a “reappreciated” trait of tumor environment driving malignancy: Perspectives in diagnosis and therapy. Cancer Metastasis Rev. 2014;33:823‐832. [DOI] [PubMed] [Google Scholar]
- 71. Damaghi M, Gillies R. Phenotypic changes of acid‐adapted cancer cells push them toward aggressiveness in their evolution in the tumor microenvironment. Cell Cycle. 2017;16:1739‐1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Raghunand N, Howison C, Sherry AD, Zhang SR, Gillies RJ. Renal and systemic pH imaging by contrast‐enhanced MRI. Magn Reson Med. 2003;49:249‐257. [DOI] [PubMed] [Google Scholar]
- 73. Wu Y, Zhang S, Soesbe TC, et al. pH imaging of mouse kidneys in vivo using a frequency‐dependent paraCEST agent. Magn Reson Med. 2016;75:2432‐2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 T1 maps for phantom #1 (A) and phantom #2 (B)
FIGURE S2 The CEST images at 4.2, 5.5, and 10 ppm for single‐slice (A,C,E) and multislice acquisition (B,D,F)
FIGURE S3 Saturation‐transfer CEST maps at 4.2 ppm and 5.5 ppm for the single‐slice (A,C) and the multislice (B,D) approach
FIGURE S4 B0 shift maps (A) and mean B0 shift region of interest–averaged values for the tumor‐bearing mouse shown in Figure 8A,B and for the kidneys of the healthy mouse shown in Figure 9C,D
TABLE S1 Coefficient of determination for the measured and calculated pH values for phantom #2
Data Availability Statement
The developed MRI‐CEST multislice sequence is freely available at https://www.cim.unito.it/website/research/research_MRI_seq.php.


