Figure 4. Integration of drug–gene interaction data to identify surrogate therapeutic targets for challenging proteins.
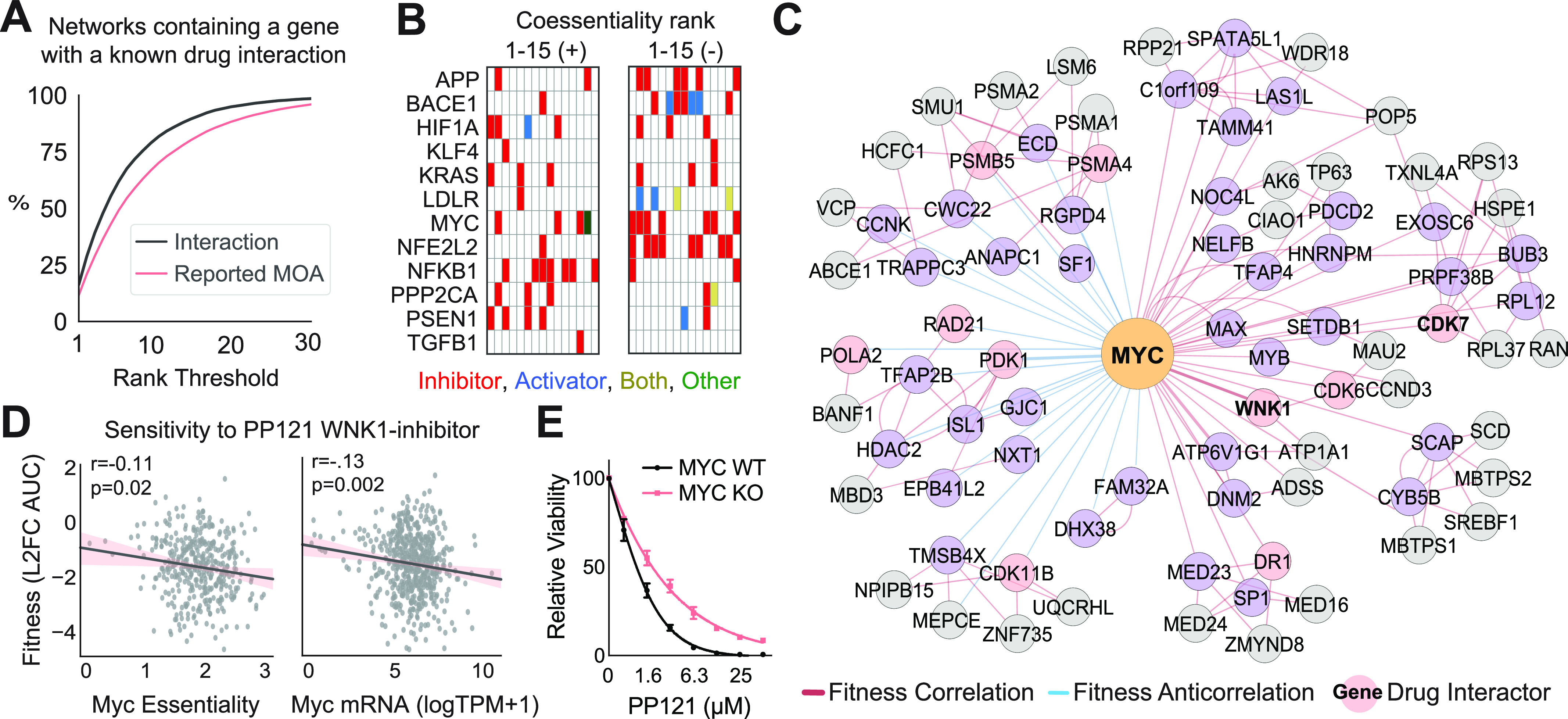
(A) Proportion of bottom-up coessentiality networks in the genome which contain at least one protein with a known gene-drug interaction in the Drug-Gene Interaction Database at the specified rank threshold. For (A), only positive primary nodes are considered. Reported mechanism of action refers to drug–gene interactions characterized with mechanisms such as “inhibitor” or “activator.” (B) Presence of drug–gene interactions with reported mechanism of action for the top 15 ranked correlations and anticorrelations for a panel of attractive therapeutic target proteins. (C) An example bottom-up network for a challenging therapeutic target, MYC, which has a coessential knockout phenotype with several genes targeted by existing drugs (red nodes). (D) Cancer cell dependence on and expression of MYC are associated with greater sensitivity to the WNK inhibitor PP121. P-value from Pearson correlation. (E) Viability of MYC KO (HO15.19) or parental MYC WT (TGR-1) rat fibroblasts treated with PP121 at the indicated concentrations. Three biological replicates per dose.
