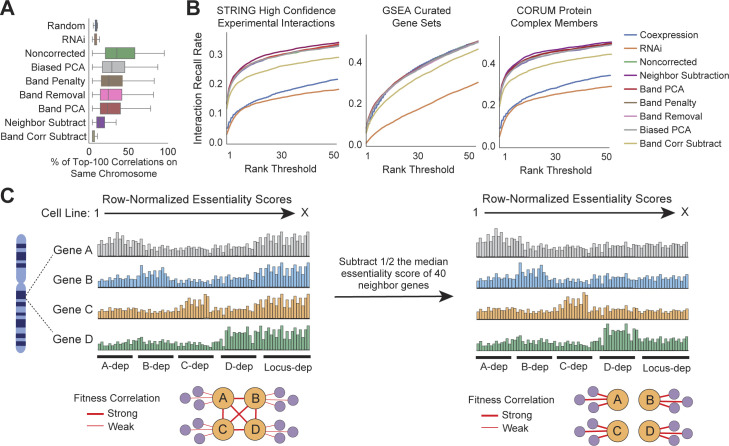
Figure S2. Benchmarking of different locus-correction approaches.
(A) All bias reduction approaches applied to the Project Achilles CRISPR-Cas9 fitness screening dataset reduce false positives (i.e., the syntenic coessentiality rate beyond expected) to different degrees. (B) The neighbor subtraction bias adjustment preprocessing approach confers the greatest performance increase for prediction of experimental interactions and protein complex membership and does not worsen the ability to recall genes in the same gene set enrichment analysis gene set. (C) A schematic illustration of the neighbor subtraction approach to reduce locus bias in CRISPR coessentiality data, where each bar represents a cell line, and the height of the bar represents dependence on the indicated gene.