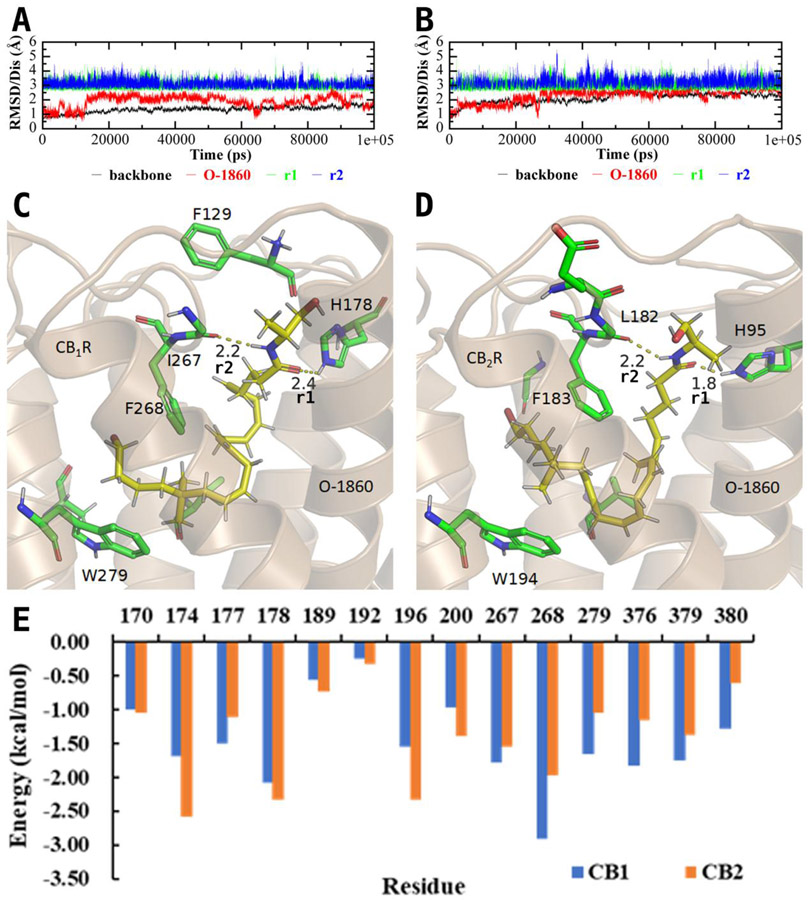
Figure 3.
(A) Room-mean-squares deviation (RMSD) of backbone atoms (black) of CB1R and heavy atoms of O-1860 (red) along with two crucial distances (r1 and r2) indicated in panel C in the MD-simulated CB1R binding with O-1860. (B) RMSD of backbone atoms (black) of CB2R and heavy atoms of O-1860 (red) along with two crucial distances (r1 and r2) indicated in panel D in the MD-simulated CB2R binding with O-1860. (C) A snapshot of the MD-simulated structure of CB1R binding with O-1860 after 100 ns. (D) A snapshot of the MD-simulated structure of CB2R binding with O-1860 after 100 ns. (E) Decomposed per-residue binding energies for residues surrounding O-1860.