Figure 1. Arouser is a substrate for endosomal microautophagy.
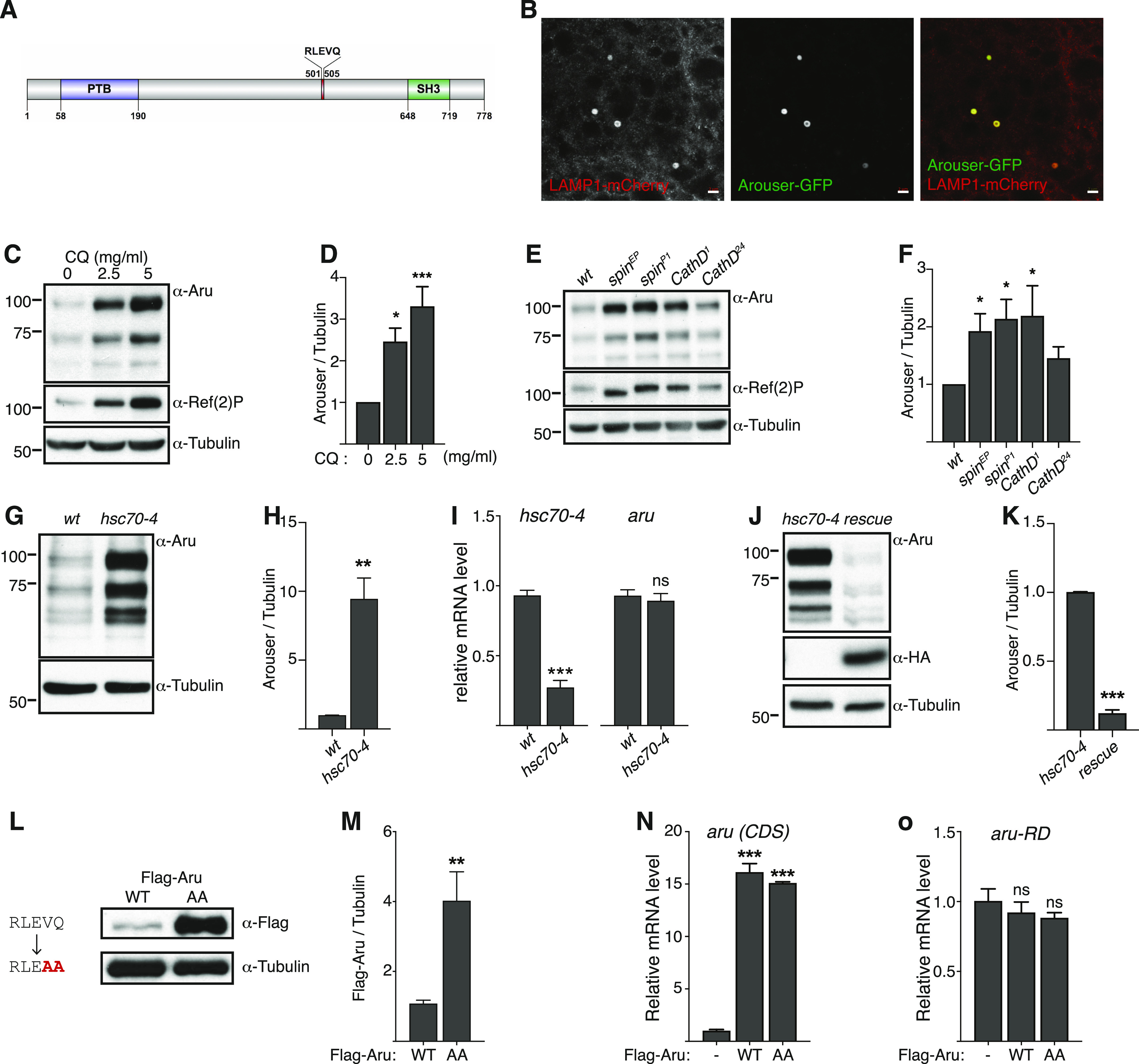
(A) Domains and motif of Arouser protein. PTB, phosphotyrosine binding domain; SH3, SRC Homology 3 Domain. (B) Airyscan confocal section of a fat body cell from starved showing the colocalisation of Arouser-GFP (green) and LAMP1-3xmCherry (red). Scale bar: 2 μm. (C, D, E, F) Western blot analysis and quantification of endogenous Arouser protein in larvae fed for 24 h with chloroquine (C, D) or in larvae with defective lysosomes (E, F). (G, H, I, J, K) Analysis of endogenous Arouser in larvae with defective eMi (G, H, I) and eMi rescue (J, K); relative gene expression levels for hsc70-4 and aru are shown in (I). (L, M) Western blot analysis and quantification of wild-type (WT) and eMi-resistant (AA) Flag-Arouser expressed in the larval fat body. (N, O) Relative gene expression for aru pan-isoform (CDS) (N) and endogenous aru using primers specific to aru-RD isoform (O) in flies expressing Flag-Aru wild-type (WT) and eMi mutant (AA); w1118 flies were used as negative control. Bar charts show means ± s.d. Statistical significance was determined using one-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001.
