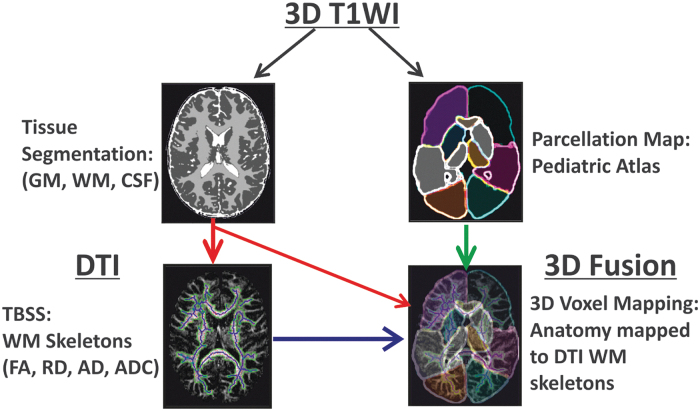
FIG. 1.
Overall scheme of image processing used in this study. The subject's high resolution T1 data were input into an atlas-based brain parcellation algorithm and segmented into tissue classes (white matter, gray matter, and cerebrospinal fluid) as well as 17 brain anatomic regions in T1/T2 space. The subject's diffusion data was co-aligned with its own T1/T2 data to transfer tissue and anatomy information to diffusion tensor imaging (DTI) spaces. Finally, tract-based statistics from DTI were fused for individual brain anatomy and tissue regions to estimate normative developmental trends in a pediatric dataset. Color image is available online.