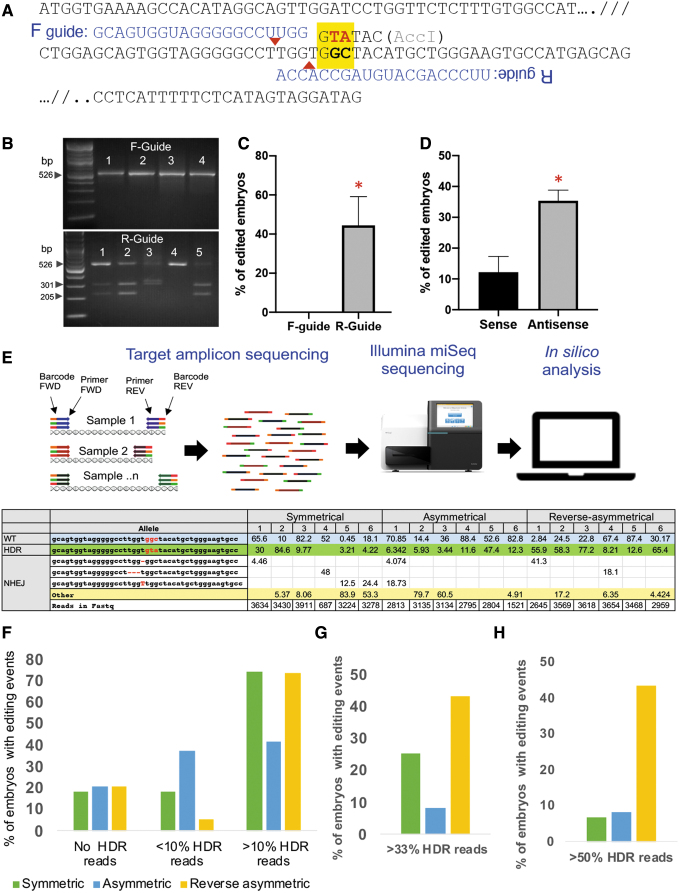
FIG. 1.
Identifying optimal CRISPR-Cas targeting reagents. (A) Schematic outlining the target site of the endogenous bovine PRNP locus. The coding sequence was truncated for convenience (indicated by “//”). The site coding for glycine (GGC) at amino acid position 127 targeted for conversion to valine (GTA) is highlighted in yellow. Successful gene targeting will result in the generation of AccI restriction enzyme site (GTATAC). Two CRISPR spacer sequences targeting the sense strand (F-guide) and antisense strand (R-guide) are shown above the target site. Putative cut-site is shown as a red triangle for each guide. (B) Representative agarose gel electrophoresis image indicating that AccI site introduction into genomic DNA results only from R-guide, and not from F-guide. Targeted genomic region was PCR amplified from embryos (numbered), amplicon fragment gel purified, AccI digested, and digestion fragments resolved on a 2% agarose gel. (C) Results from gene targeting with F- and R-guide from three triplicate experiments. No successful gene targeting was identified with F- guide (n = 14 blastocysts), whereas R-guide resulted in successful targeting with a statistically significant finding (n = 16 blastocysts; *P < 0.05). (D) Results from gene targeting with R-guide and oligos targeting either sense (n = 39 blastocysts) or antisense strand (n = 39 blastocysts). Antisense oligo resulted in better targeting efficiencies from duplicate experiments (*P < 0.05). (E) Schematic outlining Illumina miSeq targeting amplicon sequencing of blastocysts from gene targeting of embryos with various iterations of antisense oligos (sense, antisense, reverse-asymmetric). FastQ files from the miSeq run are trimmed and aligned to reference sequence. Wildtype, unmodified homology-directed repair (HDR) and nonhomologous end-joining (NHEJ) events from representative blastocysts were binned. All embryos were mosaic showing various combination of editing events. (F) The results from collating replicates over several weeks and showing editing events were shown (120 blastocysts total: 43 symmetrical, 24 asymmetrical, and 53 reverse-assymmetrical). Percentage of embryos showing no HDR reads, <10% HDR reads (low HDR frequency), and >10% reads are shown. Asymmetric oligo was least efficient among the three oligos tested. (G) Percentage of embryos showing one-third of gene targeted alleles (>33 % HDR); and (H) greater than half of the modified alleles (>50 % HDR) are shown. Reverse-asymmetric oligos resulted in better targeting efficiencies with greater number of embryos showing high HDR frequency. FWD, forward; REV, reverse.