Figure 2.
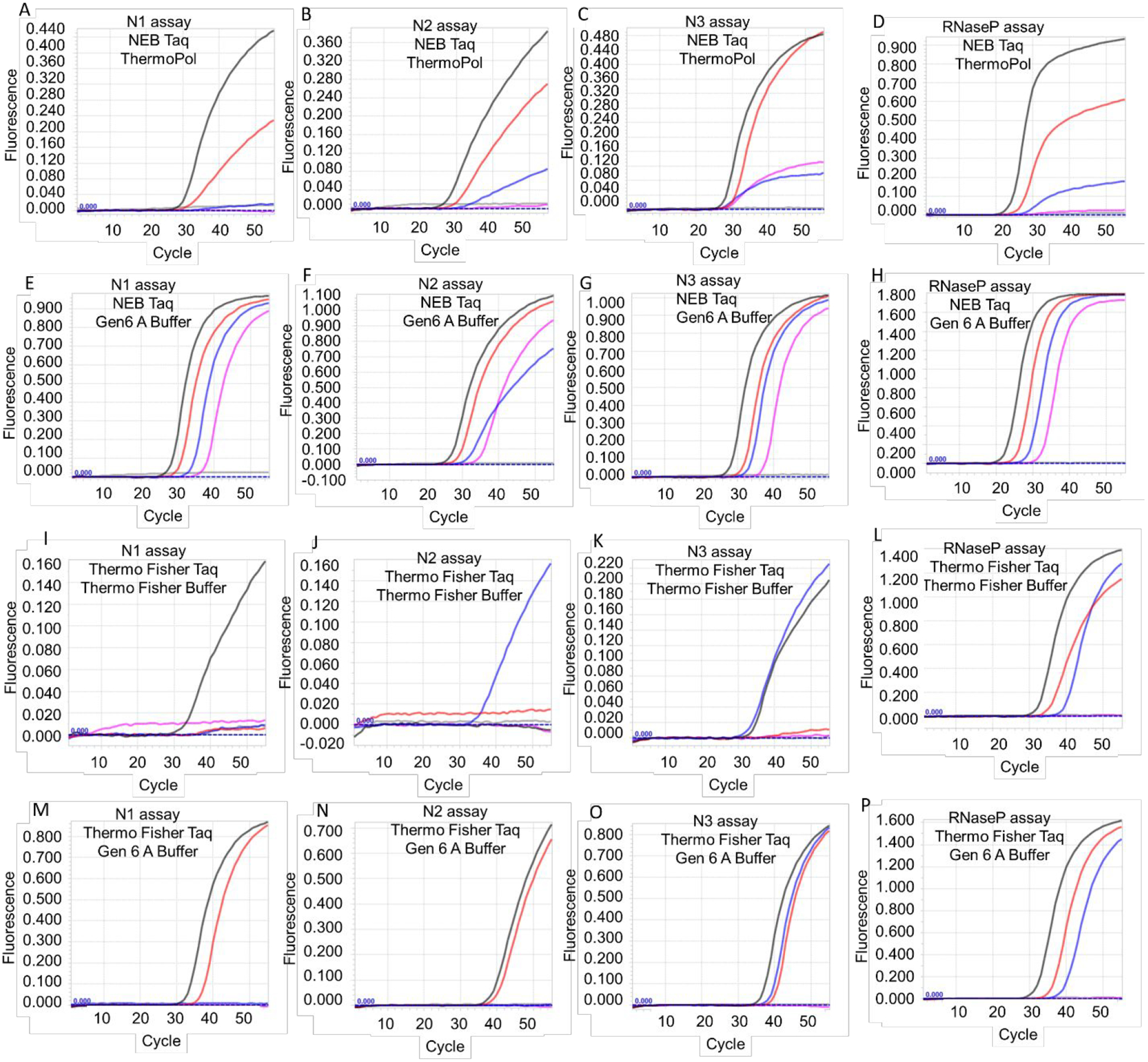
TaqMan RT-qPCR analysis of SARS-CoV-2 viral genomic RNA and RNaseP armored RNA using Taq DNA polymerase-based one-enzyme assays. CDC SARS-CoV-2 N gene assays, N1, N2, and N3, and RNaseP assay were performed using Taq DNA polymerase from either NEB (panels A-H) or Thermo Fisher (panels I-P). Assays were performed either using the companion commercial buffer (panels A-D and panels I-L) or using Gen 6 A buffer (panels E-H and panels M-P). Representative amplification curves from 6000 (black traces), 600 (red traces), 60 (blue traces), 6 (pink traces), and 0 (gray traces) copies of viral genomic RNA are depicted in panels A-C, E-G, I-K, and M-O. Representative amplification curves from 3 × 105 (black traces), 3 × 104 (red traces), 3 × 103 (blue traces), 3 × 102 (pink traces) and 0 (gray traces) copies of armored RNaseP RNA are depicted in panes D, H, L, and P.
