Figure 3: The ccp gene provides a fitness advantage for C. rodentium that is dependent on the Nox1 mouse genotype, but independent of changes in the microbiota composition.
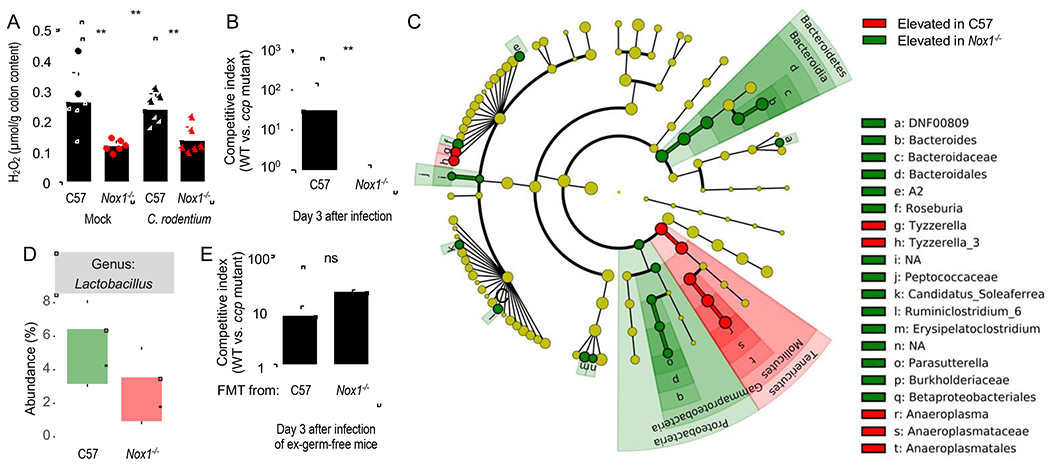
(A) C57BL/6 mice (C57, N = 6) and congenic Nox1-deficient mice (Nox1−/−, N = 6) were mock-infected or infected with C. rodentium wild type. Hydrogen peroxide was measured in the supernatant of colon contents using Amplex Red and horseradish peroxidase on day 3 after infection. (B) C57BL/6 mice (C57, N = 6) and congenic Nox1-deficient mice (Nox1−/−, N = 6) were infected with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time point. (C and D) DNA was isolated from fecal contents of C57BL/6 mice (C57, N = 6) or congenic Nox1-deficient mice (Nox1−/−, N = 6) and the microbiota composition analyzed by 16S rRNA gene amplicon sequencing. (C) Results of a linear discriminant analysis of microbiota composition in C57BL/6 mice compared to Nox1-deficient mice are displayed as a cladogram, highlighting differences in the abundance of taxa (red and green highlighting) determined by 16S rRNA gene amplicon sequencing. Taxa are arranged on a circular phylogenetic tree. Green, taxa elevated in Nox1-deficient mice compared to C57BL/6 mice; Red, taxa reduced in Nox1-deficient mice compared to C57BL/6 mice. (D) Relative abundance of Lactobacillus species. (E) Germ-free mice (Swiss Webster, N = 4) received a fecal microbiota transplant from conventional mice (C57) or congenic Nox1-deficient mice (Nox1−/−) and seven days later mice were challenged with a 1:1 mixture of the C. rodentium wild type (WT) and a ccp mutant. The competitive index (ratio of WT to mutant) in the feces was determined at the indicated time point. **, P < 0.01; ns, P > 0.05.
