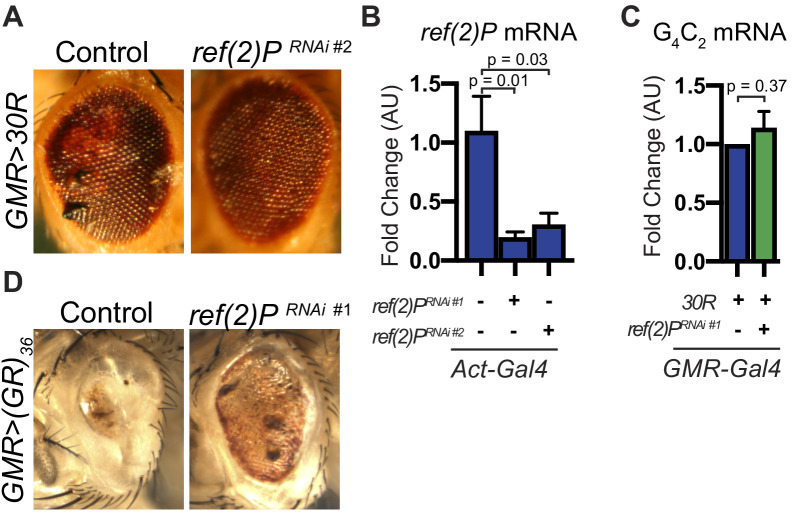
Figure 1. Autophagy receptor Ref(2)P/p62 genetically suppresses G4C2-HRE-mediated degeneration.
(A) 15-day-old Drosophila eyes expressing GMR-Gal4 +/- UAS-30R (GMR>30R) with RNAi background (control), ref(2)P RNAi#1 or overexpression (OE) of ref(2)P. (B) Quantification of external eye degeneration in A by semi-quantitative scoring system. Data are reported as mean ± SEM. Kruskal-Wallis test, p<0.0001, followed by Dunn’s multiple comparisons, n > 15 adults. (C) 15-day-old Drosophila eyes expressing GMR-Gal4 +/- UAS-36R (GMR >36R) along with UAS-luciferase RNAi (control), UAS-ref(2)P RNAi #1, UAS-ref(2)P RNAi #2, or UAS-ref(2)P OE. (D) Quantification of external eye degeneration in C by semi-quantitative scoring system. Data are reported as mean ± SEM. Kruskal-Wallis test, p<0.0001, followed by Dunn’s multiple comparisons, n = 23, 62, 28, 10 adults respectively. (E) Percent of pupal eclosion of adult flies expressing the motor neuron driver vGlut-Gal4 +/- UAS-30R and RNAi background control or UAS-ref(2)P RNAi #1. Fisher’s exact test, n > 100 pupa. (F) Percent of pupal eclosion of adult flies expressing the motor neuron driver vGlut-Gal4 +/- UAS-44R along with UAS-luciferase RNAi, UAS-ref(2)P RNAi #1, or UAS-ref(2)P OE. Fisher’s exact test, n > 55 pupa. (G) Adult Drosophila expressing UAS-30R under the control of the inducible, pan-neuronal elavGS induced with 200 µM RU486 or vehicle alone and co-expressing control or UAS-ref(2)P RNAi #1. Data are reported as mean ± SEM. One-way ANOVA, ****p<0.0001, with Sidak’s multiple comparisons test, n = 9, 8, 8, 8 groups of 10 flies.