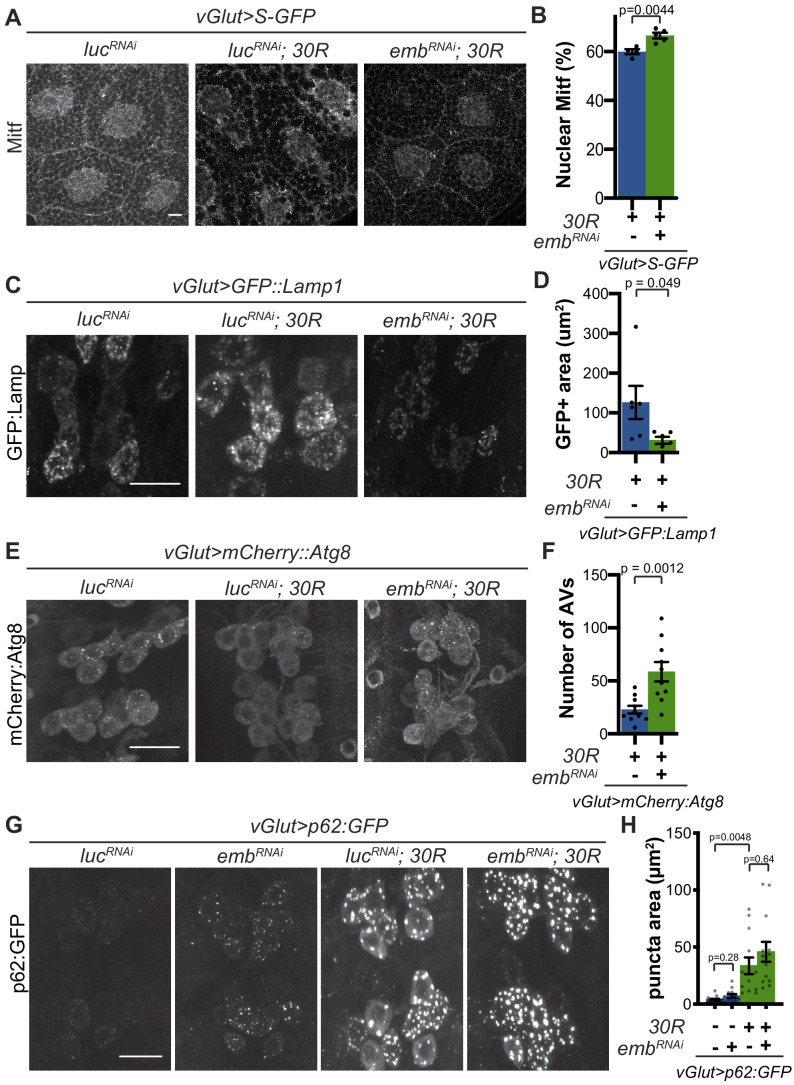
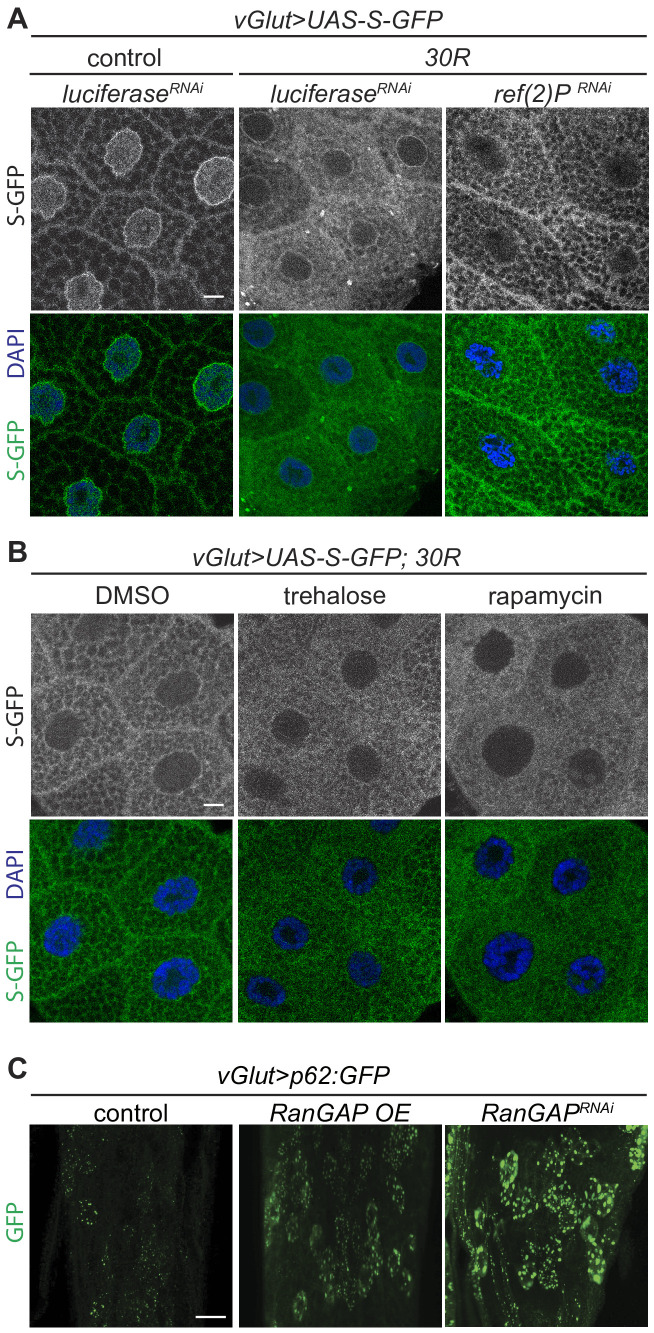
Figure 5. Modulation of nucleocytoplasmic transport rescues autophagolysosome dysfunction.
(A) Drosophila larval salivary glands stained with anti-Mitf and DAPI expressing +/- UAS-30R, UAS-shuttle-GFP (S-GFP, not shown), and either control RNAi (UAS-lucRNAi) or exportin RNAi (UAS-embRNAi ) under the control of vGlut-Gal4. Scale bar = 10 µm (B) Quantification of percent (%) nuclear Mitf in A. Data are reported as mean ± SEM. Student’s t-test, n = 4 and 5 larvae, respectively. (C) Drosophila motor neurons expressing UAS-GFP:Lamp1 (N-terminal, luminal GFP) -/+ UAS-30R and UAS-lucRNAi or exportin RNAi (UAS-embRNAi ). Scale bar = 10 µm. (D) Quantification of C. Student’s t-test, n = 6 larvae. (E) Drosophila motor neurons expressing UAS-mCherry:Atg8 +/- UAS-30R and either control RNAi (UAS-lucRNAi) or exportin RNAi (UAS-embRNAi ). Scale bar = 10 µm. (F) Quantification of E. Data are reported as mean ± SEM. Mann-Whitney test, n = 10 larvae. (G) Drosophila motor neurons expressing UAS-p62:GFP -/+ UAS-30R and either control RNAi (lucRNAi) or exportin RNAi (embRNAi ) under the control of vGlut-Gal4. Scale bar = 10 µm. (H) Quantification of G. Data are reported as mean ± SEM. Brown-Forsythe and Welch ANOVA test, p<0.0001, followed by Dunnett’s T3 multiple comparisons, n = 12–14 larvae per genotype.