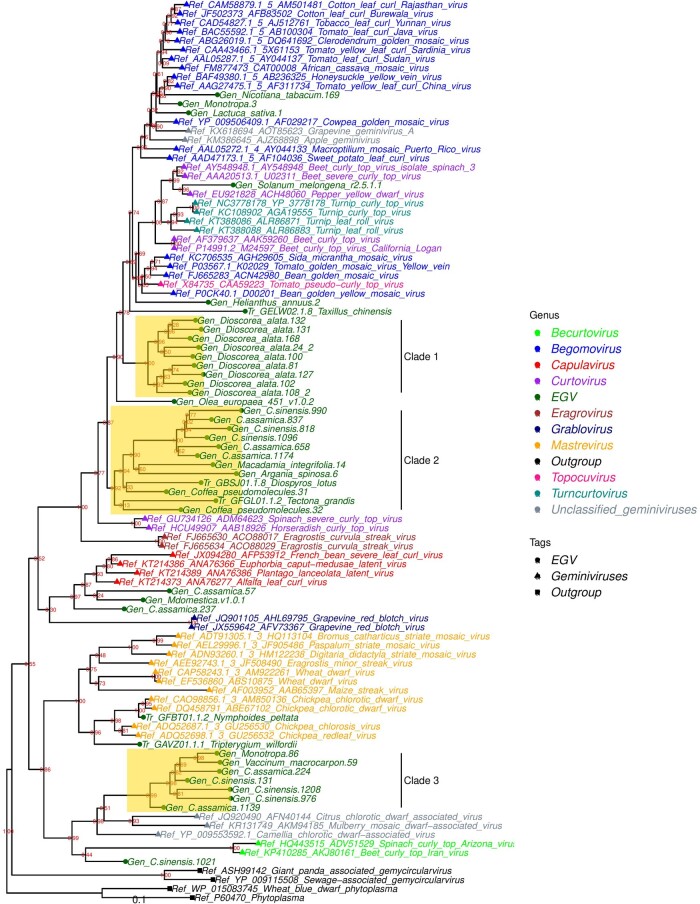
Figure 3.
Phylogenetic placement of EGV-encoded Rep proteins. Maximum-likelihood phylogenetic tree constructed using EGV Rep protein sequences retrieved from plant genome sequences (Gen; thirty-six sequences) and transcriptome datasets (Tr; 5 sequences) and from representatives of the geminivirus genus (fifty-five sequences). Distant homologues (four sequences from genomoviruses and phytoplasma) were used as outgroups. Sequences are color-coded according to virus genera, EGV, genomovirus, or phytoplasma origin. Bootstrap values are shown on the nodes of the phylogenetic tree. A scale of substitution rates is provided at the bottom of the tree.