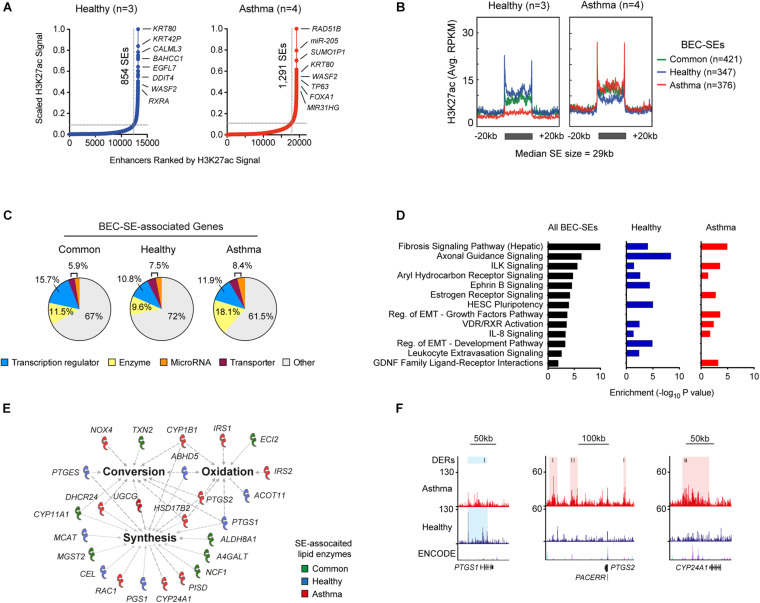
FIGURE 2.
Identification of cell-specific and asthma-associated super-enhancers in the airway epithelium. (A) Ranking of H3K27ac ChIP-Seq signal from merged Healthy (blue, n = 3) and Asthma BECs (red, n = 4) identified super-enhancers in BECs (BEC-SEs) and associated genes. BEC-SEs were shared (e.g., KRT80) and unique to healthy (e.g., CALML3) or asthma (e.g., RAD51B). (B) “Meta gene” plots summarizing H3K27ac enrichment (RPKM) in healthy and asthma BECs across Common, Healthy, and Asthma-associated SEs. (C) Consistent with super-enhancers in other cells types, BEC-SE-associated genes included transcriptional regulators (blue). However, we also identified substantial numbers of BEC-SE-associated genes encoding enzymes (yellow). (D) Pathway analysis indicating that BEC-SE-associated genes were enriched in various pathways including fibrosis, axonal development, and those implicated in epithelial biology (e.g., EMT). (E) Lipid-associated processes (bold) and enzymes encompassed by BEC-SEs. (F) Genome tracks depict H3K27ac enrichment, DERs, and location of BEC-SEs across select lipid-associated enzymes.