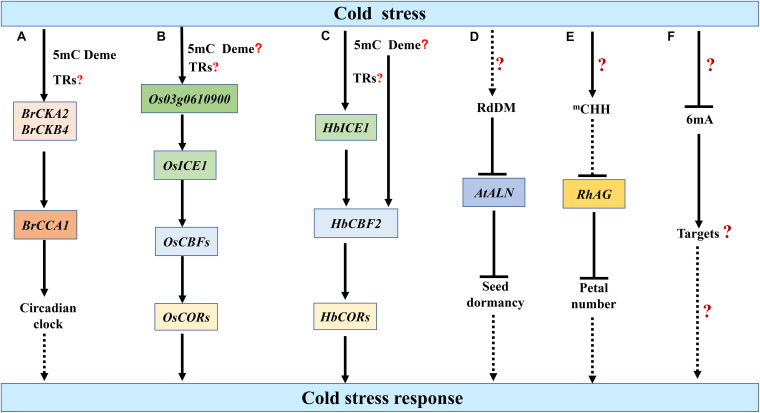
FIGURE 3.
Epigenetic regulation of plant cold stress responses by DNA methylation. (A) In Brassica rapa, prolonged cold stress mediates the demethylation and increased expression of CASEIN KINASE II A-SUBUNIT (BrCKA2) and B-SUBUNIT (BrCKB4), which may regulate cold stress response through the clock gene CIRCADIAN CLOCK ASSOCIATED 1 (BrCCA1) (Duan et al., 2017). (B) Cold stress decreases the 5mC in the promoter of Os03g0610900 and up-regulates its expression, thereby enhancing INDUCER OF CBF EXPRESSION 1 (ICE1)-mediated cold resistance (Guo et al., 2019). (C) In Hevea brasiliensis, cold stress elevates the transcriptional activities of HbICE1 as well as C-repeat binding factor 2 (HbCBF2), which may be associated with the DNA demethylation in their promoters (Tang et al., 2018). Whether cold-induced 5mC demethylation results in the up-regulation of Os03g0610900, HbICE1 and HbCBF2 under cold stress remains to be elucidated (B,C). How cold-induced 5mC demethylation integrated with the activity of transcriptional regulators (TRs) to activate downstream gene expression also needs to be investigated (A–C). (D,E) Cold stress up-regulates the 5mC levels of ALLANTOINASE (ALN) in Arabidopsis and AGAMOUS homolog RhAG in rose (Rosa hybrida). The resulting repressed expression of AtALN and RhAG may be associated with seed dormancy and petal number under cold stress, respectively (Ma N. et al., 2015; Iwasaki et al., 2019). (F) Cold stress reduces the 6mA levels in rice, but the roles of 6mA in plant responses to cold stress remain obscure (Zhang Q. et al., 2018). 5mC Deme represents the demethylation of 5mC. The speculative regulatory paths are shown with broken arrows. Many unknown targets or steps (?) remain to be investigated.