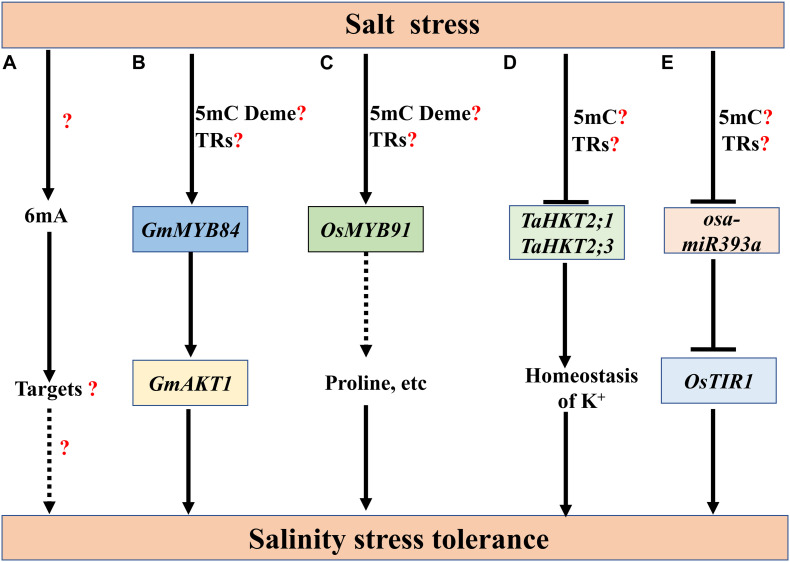
FIGURE 4.
The roles of DNA methylation in plant salt stress responses. (A) Salt stress increases the 6mA levels in rice (Zhang Q. et al., 2018), but the regulatory mechanisms underlying 6mA in salt stress remain to be investigated. (B) In soybean, salt stress induces 5mC demethylation at the promoter of MYB DOMAIN PROTEIN 84 (GmMYB84), which may be associated with its higher expression. GmMYB84 activates K+ TRANSPORTER 1 (GmAKT1) to confer salinity stress tolerance (Zhang W. et al., 2020). (C) Salt stress leads to rapid removal of 5mC from the promoter of OsMYB91, which may contribute to the salt-induced expression of OsMYB91. Plants over-expressing OsMYB91 show enhanced tolerance with significant increases of proline levels (Zhu et al., 2015). 5mC Deme represents the demethylation of 5mC. (D) Salinity stress induces an increase in 5mC levels of HIGH-AFFINITY POTASSIUM TRANSPORTER 2;1/3 (TaHKT2;1 and TaHKT2;3) that may downregulate their expression, thereby improving the salt tolerance. (E) Under salt stress, increased methylation levels of osa-miR393a promoter may lead to a lower expression of osa-miR393a, which may increase the salt tolerance through up-regulation of the target TRANSPORT INHIBITOR RESPONSE 1 (OsTIR1). How salt-induced 5mC methylation or demethylation integrated with the function of transcriptional regulators (TRs) to activate downstream gene expression remains unclear (B–E). The speculative regulatory paths are shown with broken arrows. Many unknown targets or steps (?) remain to be studied.