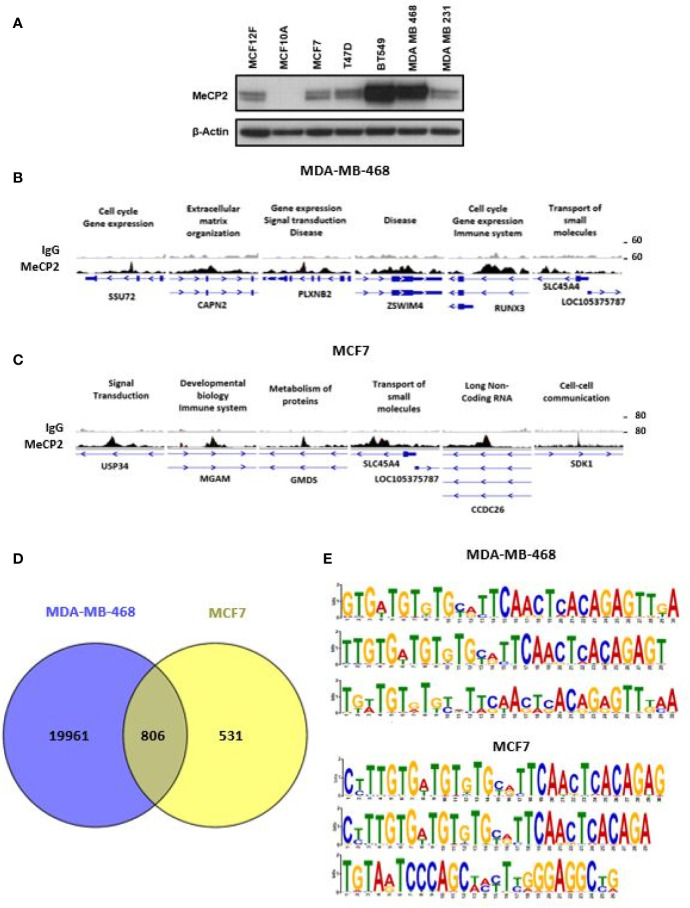
Figure 1.
ChIP-Seq identified novel MeCP2 binding motifs. (A) MeCP2 expression in breast cancer cell panel including non-cancer cell lines MCF12F and MCF10A and breast cancer cell lines MCF7, T47D, BT 549, MDA-MB-468 and MDA-MB-231. (B) An assembly of IgG (first row) and MeCP2 (second row) ChIP-Seq data in MDA-MB-468 for the SSU72, CAPN2, PLXNB2, ZSWIM4, RUNX3, and SLC45A4 genes, visualized by IGV. (C) An assembly of IgG (first row) and MeCP2 (second row) ChIP-Seq data in MCF7 for the USP34, MGAM, GMDS, SLC45A4, CCDC26 and SDK1 genes, visualized by IGV. For (B, C) each column is 4,000 bp wide. The third rows show the gene nearest to the ChIP-Seq alignment including its location, and orientation. The medium thick dark lines are the UTRs of the gene and the thicker dark regions are exons followed by thin lines with arrows which are the introns. (D) Venn diagram representing overlap of MeCP2 ChIP-Seq peaks between MDA-MB-468 and MCF7 cells. (E) MEME-ChIP (Motif Analysis of Large Nucleotide Datasets) analysis of the MeCP2 binding sites identified MeCP2 specific motifs in MDA-MB-468 cells and MCF7 cells.