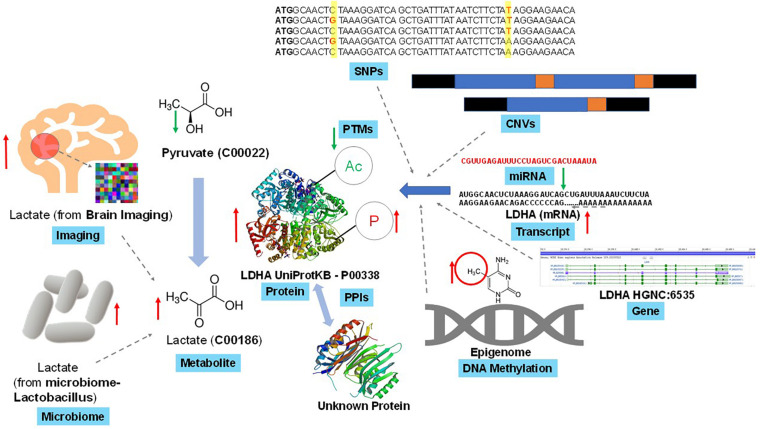
FIGURE 2.
Example of complexity and interconnectivity of omics data sources in a multi-omics framework. A simple cellular endogenous metabolite, lactate is biosynthesized enzymatically from pyruvate (another metabolite) with the help of lactate dehydrogenase (LDHA, a catalytic protein). In turn this LDHA can interact with several known and unknown proteins through protein-protein interactions to regulate its own function, and itself is subjected to diverse post-translational modifications (PTMs) that regulate its catalytic function. Lactate measurement through techniques such as in vivo brain imaging in human or other model animals can generate lactate’s spatial distribution. Gut microbiome via Lactobacillus and other microbes can synthesize lactate and release into human physiological systems to contribute to lactate levels. Lactate biosynthesis regulation can be due to various levels of genetic (e.g., SNPs, CNV, etc.), transcriptomic, post-transcriptomic (e.g., miRNA) and/or epigenetics (e.g., DNA methylation) changes on the LDHA gene. Though this is one of the well-studied set of multi-omics interactions, but one can expect more complex and unknown interactions while integrating multi-omics datasets.