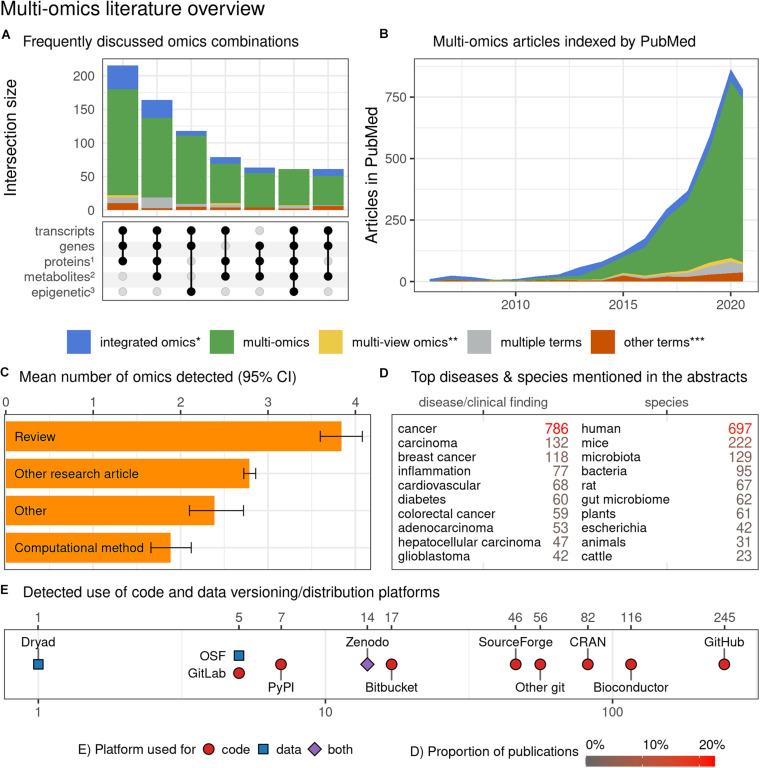
FIGURE 5.
Characterization of multi-omics literature based on a systematic screen of PubMed indexed articles (up to July 2020). (A) Combinations of omics (grouped by the characterized entities) commonly discussed occurring together in multi-omics articles (intersections with ≥ 3 omics and at least 50 papers). The proteins group (1) also includes peptides; the metabolites group (2) includes other endogenous molecules; the epigenetic group (3) encompasses all epigenetic modifications. (B) Trend plot representing the rapidly increasing number of multi-omics articles indexed in PubMed (also after adjusting for the number of articles published in matched journals – data not shown); the dip in 2020 can be attributed to indexing delay which was not accounted for in the current plot. (C) Distribution of article categories that mention different numbers of omics; while it is understandable that multi-omics “Review” category discusses many omics, the “Computational method” category articles appear to lag all other article category types. The detected number of omics may underestimate the actual numbers (due to the automated search strategy) but should put a useful lower bound on the number of omics discussed. Bootstrapped 95% confidence intervals around the mean are presented with the whiskers. (D) The number of articles mentioning the most popular clinical findings, disease terms (here screening is based on ClinVar diseases list) and species (based upon NCBI Taxonomy database). Both databases were manually filtered down to remove ambiguous terms and merge plural/singular forms. Only the abstracts were screened here. (E) The detected references to code, data versioning, distribution platforms and systems (links to repositories with deposited code/data); both the abstracts and full-texts (open-access subset, 44% of all articles) were screened. No manual curation to classify intent of the link inclusion (i.e., to share authors’ code/data vs. to report the use of a dataset/tool) was undertaken. The details of the methods with reproducible code are available at github.com/krassowski/multi-omics-state-of-the-field. The comprehensive search terms (see the online repository for details) were collapsed into four categories; integrated omics (*) includes integromics and integrative omics, multi-view (**) includes multi-view| block| source| modal omics, other terms (***) include pan-, trans-, poly-, cross-omics.