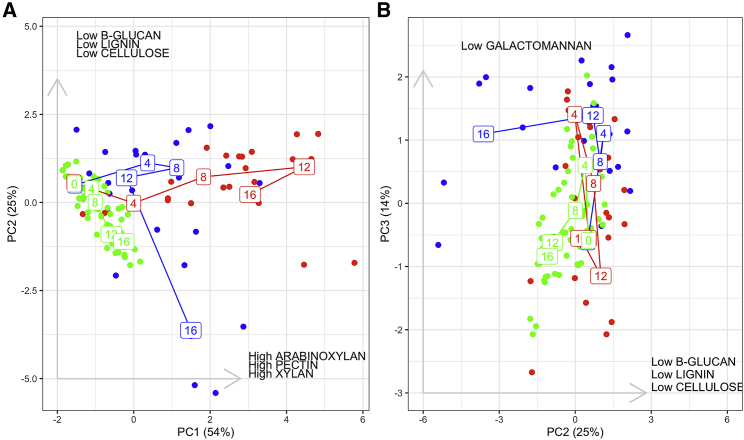
Figure 1.
Trajectories of Stenotrophomonas sp. Metabolic Phenotype Evolution
Ordination plots from a principal components analysis (PCA) of the Stenotrophomonas sp. metabolic phenotype over time. The first 3 principal components (PC) captured 92% of the variation in substrate use, and thus, these PCs were plotted to visualize the evolutionary trajectories of our treatments. Plots show (A) PC1 (54%) against PC2 (25%) and (B) PC2 against PC3 (14%). The variation in resource use associated with each PC is stated on each axis. Lines show evolutionary trajectories for the Stenotrophomonas sp. metabolic phenotype in the monoculture (MC; green), reset polyculture (RP; red), and dynamic polyculture (DP; blue) treatments; dots show values for each individual replicate over time (denoted by transfer number labels on each line). Use of individual substrates over time are plotted in Figure S1. Individual replicate trajectories for the DP treatment are plotted in Figure S2. The raw data is provided in Data S1.