Figure 6.
Formaldehyde-Accumulating Aldh2−/−Adh5−/− Mice Reveal a Mutation Signature
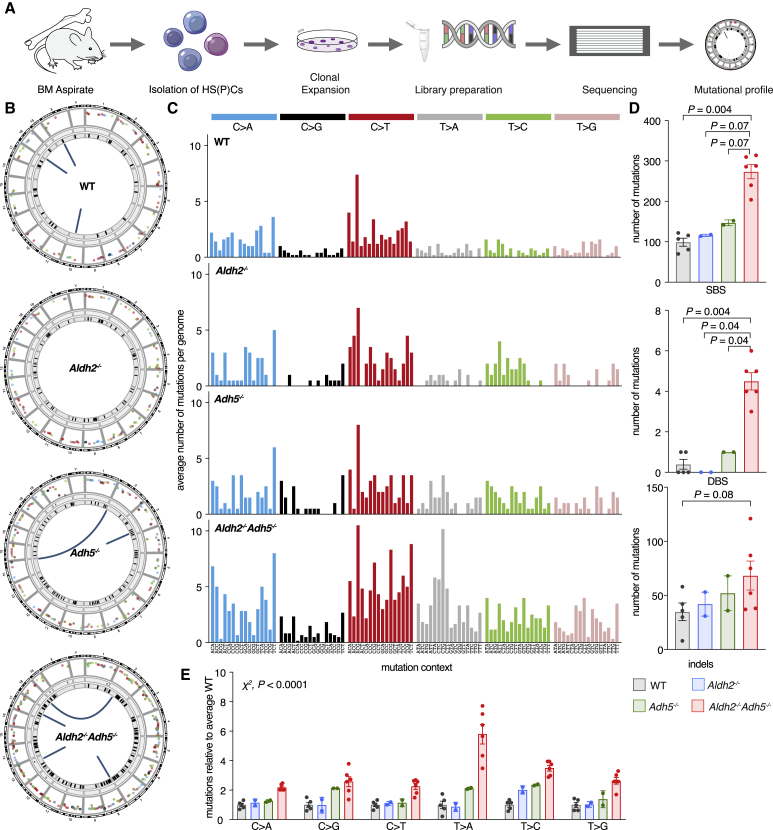
(A) Whole-genome sequencing of HSPCs.
(B) Circos plots highlighting the different types and levels of mutations from a representative Aldh2−/−Adh5−/− mouse and controls. The outermost ring represents each chromosome, followed by sequential rings highlighting single-base substitutions (SBSs) as a rainfall plot (color-coding of substitution types as in C), tandem base substitutions (DBSs), and insertions or deletions (indels). Chromosomal rearrangements are represented by lines linking the translocated chromosomes at the center.
(C) Aggregated mutational profile of SBSs in HSPC genomes. Each mutation is assigned to the pyrimidine base of the originating base pair; within each of the 6 main mutation types, the sequence context of 5′ and 3′ flanking bases is shown in alphabetical order.
(D) Frequency of SBSs, DBSs, and indels (mean ± SEM; number of HSPC genomes analyzed = 5, 2, 2, 6 from left to right; two-tailed Mann-Whitney U test).
(E) Relative mutation number at each base, normalized to the average HSPC clone from WT litter-matched 40-week-old animals (mean ± SEM; n = 5, 2, 2, 6 from left to right; χ2 test comparing the aggregate number of mutations of each type between the WT and Aldh2−/−Adh5−/−).
See also Figure S6.