Abstract
Background:
Intratumor heterogeneity (ITH) has been shown to be inversely associated with immune infiltration in several cancers including clear cell renal cell carcinoma (ccRCC), but it remains unclear whether ITH is associated with response to immunotherapy (e.g. PD-1 blockade) in ccRCC.
Methods:
We quantified ITH using mutant-allele tumor heterogeneity, investigated the association of ITH with immune parameters in patients with ccRCC (n = 336) as well as those with papillary RCC (pRCC, n = 280) from The Cancer Genome Atlas, and validations were conducted in patients with ccRCC from an independent cohort (n = 152). The relationship between ITH and response to anti-PD-1 immunotherapy was explored in patients with metastatic ccRCC from a clinical trial of anti-PD-1 therapy (n = 35), and validated in three equal-size simulated data sets (n = 60) generated by random sampling with replacement based on this clinical trial cohort.
Results:
In ccRCC, low ITH was associated with better survival, more reductions in tumor burden, and clinical benefit of anti-PD-1 immunotherapy through modulating immune activity involving more neoantigens, elevated expression of HLA class I genes, and higher abundance of dendritic cells. Furthermore, we found that the association between the level of ITH and response to PD-1 blockade was independent of the mutation status of PBRM1 and that integrating both factors performed better than the individual predictors in predicting the benefit of anti-PD-1 immunotherapy in ccRCC patients. In pRCC, increased immune activity was also observed in low- versus high-ITH tumors, including higher neoantigen counts, increased abundance of monocytes, and decreased expression of PD-L1 and PD-L2.
Conclusions:
ITH may be helpful in the identification of patients who could benefit from PD-1 blockade in ccRCC, and even in pRCC where no genomic metrics has been found to correlate with response to immune checkpoint inhibitors.
Keywords: anti-PD-1 immunotherapy, immune cell infiltration, intratumor heterogeneity, neoantigen, renal cell carcinoma
Introduction
Intratumor heterogeneity (ITH), also known as clonal heterogeneity, refers to the genetic diversity of subclones within a single tumor. With the clinical application of next-generation sequencing technology, computational methods (e.g. ABSOLUTE,1 PyClone,2 and EXPANDS3) have been developed to quantify ITH based on information such as copy number alterations, variant allele frequencies, and tumor purity from whole-exome sequencing data and explore its clinical relevance in cancers. With the aid of these tools, high ITH has been reported to correlate with worse clinical outcome.4,5 Recently, an inverse association was revealed between ITH and immune infiltration in a pan-cancer study, with the association being most significant in clear cell renal cell carcinoma (ccRCC).5 Furthermore, low ITH was shown to correlate with stronger T-cell immunoreactivity and sensitivity to immune checkpoint therapy (ICT) in melanoma and non-small-cell lung cancer (NSCLC) from the perspective of clonal versus subclonal neoantigens.6 Moreover, Wolf et al. recently demonstrated in melanoma that low ITH was associated with better survival in patients who were or were not treated with ICT, potentially due to elevated T-cell reactivity and tumor infiltration.7 However, it remains unclear whether ITH is associated with the clinical benefit of ICT in ccRCC, despite the inverse association of ITH with immune cell infiltration and clinical outcome that has been observed in ccRCC.5,8
Renal cell carcinoma (RCC), the seventh most frequently diagnosed malignancy,8 is a heterogeneous group of cancers with significant ITH.9 Of the multiple subtypes in RCC, ccRCC is the most common subtype, accounting for 70–75%,10 while papillary RCC (pRCC) makes up the majority of the remaining cases with a prevalence of 15–20%.11 In recent decades, the treatment of RCC has evolved from nonspecific immunotherapy to targeted therapy, and now to novel immunotherapy such as ICT.10 Although 25% of patients treated with nivolumab can experience durable response, the positive relationship between nonsynonymous mutation load and clinical benefit of ICT, which exists in other tumors where ICT has been successful, such as melanoma and NSCLC, has not been observed in RCC.12,13 Recently, loss-of-function (LOF) mutations in the PBRM1 gene were found to be associated with clinical benefit of ICT in ccRCC.14,15 However, some ccRCC patients who did not have LOF mutations in the PBRM1 gene could still benefit from the treatment of ICT, implying that there might be other genomic metrics which were associated with the clinical benefit of ICT in ccRCC. Additionally, it remains largely unknown which genomic features are correlated with response to ICT in pRCC where PBRM1 is rarely mutated. Therefore, it is imperative to examine whether ITH is correlated with clinical benefit of ICT in two most common RCC subtypes—ccRCC and pRCC.
Mutant-allele tumor heterogeneity (MATH)16 is an ITH quantitative method which quantifies ITH based on the distribution of allele frequencies of somatic mutations from whole-exome sequencing data. As a fast and simple ITH quantitative method, the inverse association of ITH with clinical outcome17–20 and immune infiltration21–23 has also been revealed using MATH.16 Here, by quantifying ITH with MATH,16 we examined the association of ITH with immune parameters and clinical benefit of anti-programmed cell death-1 receptor (anti-PD-1) immunotherapy in patients with RCC. We found that low ITH was associated with elevated immune activity and increased response to PD-1 blockade in RCC. Furthermore, distinct mechanisms might be involved in tumors from different RCC subtypes to escape from immune surveillance. In addition, ITH seems to act as a biomarker in a non-RCC-specific manner in influencing the clinical benefit of anti-PD-1 immunotherapy, and MATH could be a fast and accurate measurement for ITH in RCC. These relationships may yield insights into how ITH might affect patient survival and cancer therapy, and help to develop predictive biomarkers for response to ICT.
Methods
Data collection and preprocessing
To explore the relationship between ITH and immune parameters in RCC, somatic mutations, FPKM-normalized gene expressions, and clinical information for the patients with ccRCC (n = 336) and pRCC (n = 280) were obtained from The Cancer Genome Atlas (TCGA) public access portal. As TPM-normalized gene expression data were recommended for immune cell deconvolution,24 gene expression data were then transformed from FPKM-normalized to TPM-normalized using the following formula: TPM = exp[log(FPKM) – log(sum(FPKM)) + log(1e6)] according to Pachter.25 In addition, somatic mutation data and clinical information for patients with metastatic ccRCC on a clinical trial of anti-PD-1 therapy (n = 35) were obtained from a published study,14 so as to evaluate the association between ITH and clinical benefit of PD-1 blockade. False positive somatic mutations were filtered for the above data sets according to the criteria in Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT).26 Specifically, variants were filtered using the following criteria: coverage depth ⩾20×, mutant reads ⩾8, and variant frequency ⩾2%.
Patients with ccRCC from Wang et al.27 (n = 152) served as the validation cohort for the TCGA data sets. The whole-exome and transcriptome data for the cohort were downloaded from NCBI Sequence Read Archive. RNA reads were aligned to the hg38 genome assembly using STAR28 and quantified with RSEM.29 DNA reads were analyzed as previously described,27 except that somatic mutations were called using Strelka230 and annotated with ANNOVAR.31 The strict filtration criteria used by Wang et al.27 to get reliable somatic mutations was also applied in this study. In addition, three equal-size simulated data sets (n = 60) were generated through random sampling with replacement based on the samples from the clinical trial of anti-PD-1 therapy in ccRCC14 to serve as the validation cohorts for the clinical trial cohort.
Ethic approval and informed consent were not sought for the present study given that all the data used in this study were obtained from public data sets and could not be individually identifiable.
ITH quantification
MATH score,16 which quantifies the dispersion of allele frequencies of somatic mutations from whole-exome sequencing data, was used to measure ITH for the samples included in this study. As MATH scores were previously reported to vary with differences in sequencing strategies (i.e. different sequencing methods and differences in sequencing depth) and baseline characteristics between the data sets,20 the function surv_cutpoint from R package survminer was applied to determine the optimal MATH score based on survival information of the patients to distinguish high- from low-ITH tumors for each data set.
To ensure that the MATH score can be an accurate reflection of ITH, subclonal genome fractions determined by ABSOLUTE1 were obtained from a published study by TCGA group32 for the tumors with pRCC or ccRCC from TCGA. As for the ccRCC tumors from Wang et al.,27 subclonal genome fractions were determined by ABSOLUTE1 based on segmentation data from CNVkit33 and somatic point mutations identified above.
Neoantigen prediction
Neoantigen count data for the tumors with pRCC or ccRCC from TCGA were obtained from a published study from the TCGA group.32 As for the ccRCC tumors from Wang et al.,27 potential neoantigenic peptides were identified using NeoPredPipe,34 based on human lymphocyte antigen (HLA) types calls from OptiType,35 and only those with strong binding affinity according to NeoPrePipe34 were retained for downstream analysis.
Immune cell deconvolution
The evaluation of tumor-infiltrating immune cells was performed using four different tools (i.e. MCPCounter,36 quanTIseq,37 CIBERSORT,38 and ImmuCellAI39) for the tumors from TCGA and Wang et al.,27 and a type of immune cell would be considered differentially distributed in high- versus low-ITH tumors when the same trend was observed in the results of at least two tools. Specifically, based on TPM-normalized gene expression data, estimation for immune cell abundance were performed using MCPCounter36 and quanTIseq37 through the R package immunedeconv24 and using ImmuCellAI39 through its webserver(http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/analysis) with default signature files. Additionally, CIBERSORT38 was run with the default signature matrix at 1000 permutations, and immune cell fraction in absolute mode was finally obtained for the samples that met the requirements of CIBERSORT p-value <0.05, resulting in a total of 163, 245 and 82 samples for pRCC, ccRCC from TCGA and Wang et al.,27 respectively. In order to compare with results from other tools, results from CIBERSORT38 were further processed as follows: B.cells = B.cells.naive + B.cells.memory; T.cells.CD4 = T.cells.CD4.naive + T.cells.CD4.memory.resting + T.cells.CD4.memory.activated; NK.cells = NK.cells.resting + NK.cells.activated; Macrophages = Macrophages.M0 + Macrophages.M1 + Macrophages.M2; Dendritic.cells = Dendritic.cells.resting + Dendritic.cells.activated; Mast.cells = Mast.cells.resting + Mast.cells.activated.
Statistical analysis
The Wilcoxon rank-sum test was used to compare subclonal genome fractions, neoantigen counts, immune cell infiltration, immune gene expression, and maximum change in tumor burden in high- versus low-ITH tumors. The association of ITH with the clinical benefit of immunotherapy and mutation status of the PBRM1 gene was assessed using the Fisher’s exact test or the Chi-square test, while the correlation between ITH and subclonal genome fraction determined by ABSOLUTE1 was evaluated with Pearson’s correlation analyses. The Kaplan–Meier method was used to estimate the distribution of survival times, and the log-rank test was used to compare distributions. Univariate and multivariate Cox proportional hazard regression models were adopted to determine hazard ratios (HRs) and 95% confidence intervals (CIs). All the survival analyses were performed in R (version 3.5) using survival package. All p-values were two-sided, and p-values less than 0.05 were considered statistically significant. All statistical analyses were performed using R (version 3.5).
Results
MATH score separates low from high ITH in RCC
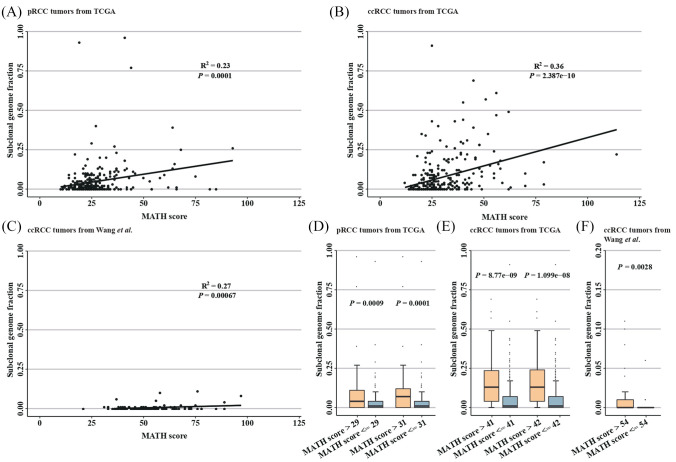
MATH score,16 a reflection of the dispersion of variant allele frequencies in tumors, ranged from 12 to 114 with a median of 26 in ccRCC patients from TCGA, from 21 to 97 with a median of 50.5 in ccRCC patients from Wang et al.,27 from 16 to 63 with a median of 34 in patients with metastatic ccRCC from the clinical trial of anti-PD-1 therapy,14 and from 10 to 93 with a median of 24 in pRCC patients from TCGA (Table1 and Figure S1). Notably, a positive correlation was observed between MATH score and subclonal genome fraction determined by ABSOLUTE (1), whether in pRCC (R2 = 0.23, p = 0.0001, Figure 1A), ccRCC samples from TCGA (R2 = 0.36, p = 2.387e-10; Figure 1B), or those from Wang et al.27 (R2 = 0.27, p = 0.00067; Figure 1C).
Table 1.
Characteristics of the patients included in this study.
| Patients | TCGA |
Miao et al.14 |
Wang et al.27 |
|
|---|---|---|---|---|
| ccRCC (n = 336) | pRCC (n = 280) | ccRCC (n = 35) | ccRCC (n = 152) | |
| Median MATH score (range) | 26 (12–114) | 24 (10–93) | 34 (16–63) | 50.5 (21–97) |
| Cutpoint of MATH score | 41 | 31 | 42 | NA |
| ITH, n | ||||
| Low | 286 | 219 | 24 | NA |
| High | 50 | 61 | 11 | |
| Median age, yr (range) | 60 (26–90) | 61 (28–88) | 63 (36–77) | 55.5 (25–87) |
| Gender, n | ||||
| Female | 120 | 76 | 13 | 41 |
| Male | 216 | 204 | 22 | 111 |
| Race, n | ||||
| White | 272 | 199 | NA | 0 |
| Black or African American | 52 | 59 | 0 | |
| Asian | 6 | 6 | 152 | |
| American Indian or Alaska native | 0 | 2 | 0 | |
| Unspecified | 6 | 14 | 0 | |
| Tumor stage, n | ||||
| Stage I | 192 | 168 | 0 | 94 |
| Stage II | 33 | 21 | 0 | 3 |
| Stage III | 68 | 49 | 0 | 41 |
| Stage IV | 41 | 15 | 35 | 14 |
| Unspecified | 2 | 27 | 0 | 0 |
| Metastatic spread, n | ||||
| M0 | 267 | 94 | 0 | 142 |
| M1 | 37 | 9 | 35 | 10 |
| MX | 30 | 162 | 0 | 0 |
| Unspecified | 2 | 15 | 0 | 0 |
| Histological subtype, n | ||||
| Type 1 | NA | 72 | NA | NA |
| Type 2 | 82 | |||
| Unspecified | 126 | |||
| Tumor grade, n | ||||
| G1 | 13 | NA | NA | NA |
| G2 | 150 | |||
| G3 | 123 | |||
| G4 | 43 | |||
| GX | 4 | |||
| Unspecified | 3 | |||
| Response to nivolumab | ||||
| CB | NA | NA | 11 | NA |
| IB | 11 | |||
| NCB | 13 | |||
CB, clinical benefit; ccRCC, clear cell renal cell carcinoma; IB, intermediate benefit; ITH, intratumor heterogeneity; MATH, mutant-allele tumor heterogeneity; NCB, no clinical benefit; pRCC, papillary renal cell carcinoma.
Figure 1.
MATH score separates low from high ITH in RCC.
Association between MATH score and subclonal genome fraction determined by ABSOLUTE (1) in pRCC tumors (A), ccRCC tumors from TCGA (B) and Wang et al. (C). R2 and p-values are from Pearson’s correlation analyses. (D) The cutpoints of MATH scores based on the survival information (PFS and DSS, respectively) of the patients with pRCC from TCGA, and a MATH score of 31 achieved a stronger significance in distinguishing high from low subclonal genome fraction. (E) The cutpoints of MATH scores based on survival information (i.e. PFS) of the patients with ccRCC from TCGA41 and those from the clinical trial of anti-PD-1 therapy,42 and a MATH score of 41 performed better in differentiating high from low subclonal genome fraction for ccRCC tumors from TCGA. (F) The MATH score that best distinguish high from low subclonal genome fraction in ccRCC tumors from Wang et al.
ccRCC, clear cell renal cell carcinoma; DSS, disease-specific survival; ITH, intratumor heterogeneity; MATH, mutant-allele tumor heterogeneity; pRCC, papillary renal cell carcinoma; PFS, progression-free survival; RCC, renal cell carcinoma; TCGA, The Cancer Genome Atlas.
In pRCC patients, MATH scores of 31 and 29 were obtained after MATH cutpoint determination based on disease-specific survival (DSS) and progression-free survival (PFS), respectively. To determine which cutpoint could better differentiate high- from low-ITH tumors, we examined subclonal genome fractions in high- versus low-ITH tumors. Although high-ITH tumors showed significantly larger proportion of subclonal mutations compared with low-ITH tumors using both cutpoints, the MATH score of 31 achieved a stronger significance (p = 0.0001 for 31; p = 0.0009 for 29; Figure 1D). Therefore, 219 (78%) patients with pRCC were categorized as low-ITH tumors (MATH ⩽31), and 3% (6/219) of these patients had distant metastasis. In patients with ccRCC from TCGA, a MATH score of 41 was obtained after MATH cutpoint determination based on either DSS or PFS. A total of 286 (85%) patients were thus classified as low-ITH tumors, among which 9% (27/286) had distant metastasis. Additionally, MATH cutpoint determination based on the PFS data (DSS data were not available) resulted in a MATH score of 42 for the patients with metastatic ccRCC from the clinical trial of anti-PD-1 therapy, and 24 patients were classified as low-ITH tumors. Please see Table S1 for the MATH score and the level of ITH for each patient.
Since the MATH cutpoint showed a little difference between patients with ccRCC from TCGA and those with metastatic ccRCC from the clinical trial, we next performed ITH classification in ccRCC tumors from TCGA using the cutpoint determined for metastatic ccRCC patients from the trial, and examined subclonal genome fractions in high- and low-ITH tumors. A significantly larger proportion of subclonal mutations was observed in high- versus low-ITH tumors, but the association was less significant (p = 1.099e-08 for 42; p = 8.77e-09 for 41; Figure 1E), suggesting that the MATH cutpoint should be determined for each data set to better differentiate high- from low-ITH tumors. Due to the lack of survival information, MATH cutpoint determination was not performed for the ccRCC tumors from Wang et al.,27 but a MATH score of 54 was found to best differentiate high from low subclonal genome fraction (p = 0.0028; Figure 1F).
Low ITH correlates with increased immune activity in ccRCC patients
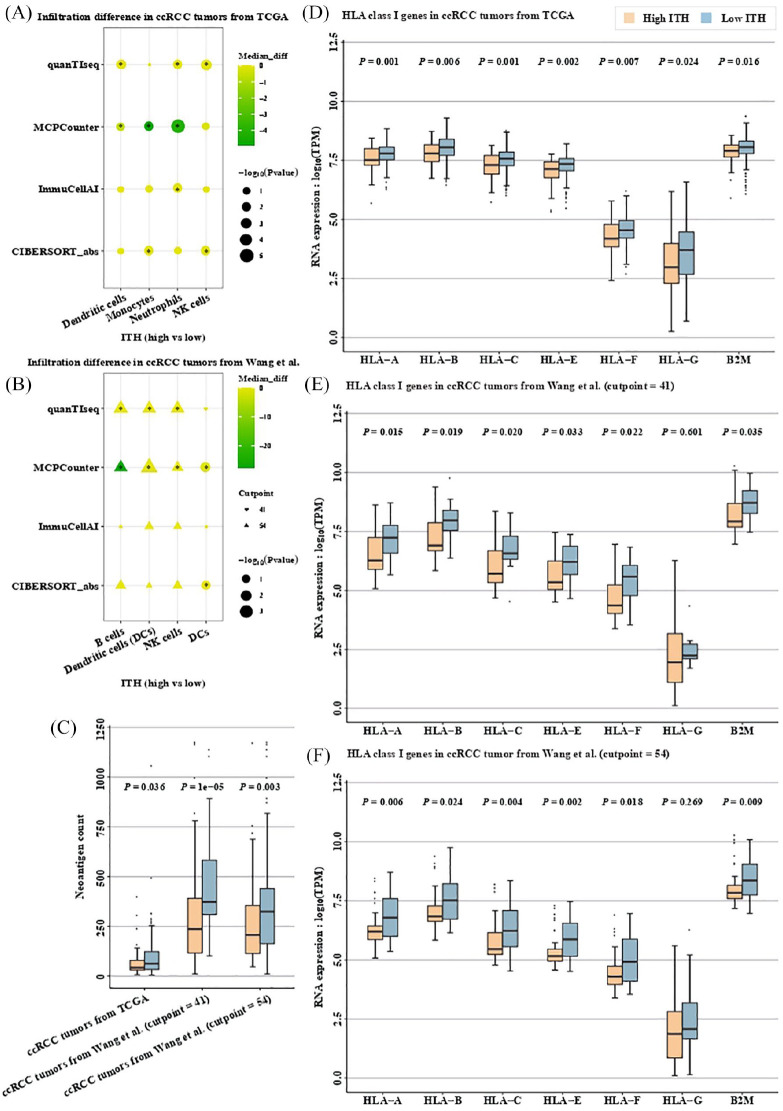
Several years ago, an inverse association was revealed between ITH and immune cell infiltration in ccRCC.5 Earlier this year, dendritic cells (DCs) were shown to be associated with response to PD-L1 blockade in RCC and NSCLC41 Therefore, we first examined immune cell infiltration between high- and low-ITH tumors in patients with ccRCC. Interestingly, in ccRCC tumors from TCGA, several types of immune cells including monocytes, DCs, natural killer (NK) cells and neutrophils showed higher abundance in low- versus high-ITH tumors (monocytes: p = 0.0177 by MCPCounter and p = 0.0154 by CIBERSORT; DCs: p = 0.0478 by MCPCounter and p = 0.0087 by quanTIseq; NK cells: p = 0.0013 by quanTIseq and p = 0.0177 by CIBERSORT; neutrophils: p = 0.00003 by MCPCounter, p = 0.0169 by quanTIseq and p = 0.025 by ImmuCellAI; Figure 2A). Higher abundance of DCs in low- versus high-ITH tumors was also observed in ccRCC patients from Wang et al.27 when a MATH score of 41 was used to differentiate high- from low-ITH tumors (p = 0.01 by MCPCounter and p = 0.03 by CIBERSORT; Figure 2B). When a MATH score of 54 was applied, not only DCs, but NK cells and B cells showed higher abundance in low- versus high-ITH tumors (DCs: p = 0.0006 by MCPCounter and p = 0.01 by quanTIseq; NK cells: p = 0.04 by MCPCounter and p = 0.011 by quanTIseq; B cells: p = 0.013 by MCPCounter and p = 0.006 by quanTIseq; Figure 2C). As for the immune cells unmentioned above, no significant difference was observed between high- versus low-ITH tumors in the results of at least two tools (Figure S2). Notably, DCs are the most potent antigen-presenting cells that process and present antigens to T cells.42
Figure 2.
Low ITH correlates with increased immune activity in ccRCC.
Infiltration difference between high- versus low-ITH tumors in patients with ccRCC from TCGA (A) and Wang et al. (B). *denotes p < 0.05. (C) Neoantigen counts derived from both SNVs and InDels between high- versus low-ITH tumors in patients with ccRCC. RNA expression of HLA class I genes in high- versus low-ITH tumors from ccRCC patients from TCGA (D), and Wang et al. using a MATH score of 41 (E) or 54 (F) as cutpoint. In ccRCC tumors from TCGA, tumors with MATH score ⩽41 were classified as low ITH, and high ITH otherwise. In ccRCC tumors from Wang et al., both MATH scores of 41 and 54 (the MATH score that best different high from low subclonal genome fraction in ccRCC tumors from Wang et al.) were used to perform ITH classification.
ccRCC, clear cell renal cell carcinoma; HLA, human lymphocyte antigen; ITH, intratumor heterogeneity; MATH, mutant-allele tumor heterogeneity; PFS, progression-free survival; SNV, single nucleotide variation; TCGA, The Cancer Genome Atlas.
Recently, RCC was reported to have the highest pan-cancer number of small insertions and deletions (InDels) that could generate more neoantigens (i.e. antigens derived from tumor-specific mutated genes) than nonsynonymous single nucleotide variations (SNVs).43 Therefore, neoantigen counts derived from both SNVs (neoAgs_SNVs) and InDels (neoAgs_InDels) were next examined between high- and low-ITH tumors. In patients with ccRCC from TCGA, the combined neoantigen counts were significantly higher in low- versus high-ITH tumors (p = 0.036; Figure 2D), and similar trend was observed in neoAgs_InDels (p = 0.01; Figure S3A) but not neoAgs_SNVs (p = 0.34; Figure S3B). In patients with ccRCC from Wang et al.,27 low-ITH tumors also show significantly higher combined neoantigen counts whether a MATH score of 41 (p = 1e-05; Figure 2D) or 54 (p = 0.003; Figure 2D) was used to differentiate high- from low-ITH tumors, yet similar trend was observed in neoAgs_SNVs (p = 5.7e-08 when the cutpoint of MATH score was 41 and p = 5e-05 when the cutpoint of MATH score was 54; Figure S3B) rather than neoAgs_InDels (p = 0.076 when the cutpoint of MATH score was 41 and p = 0.381 when the cutpoint of MATH score was 54; Figure S3A).
Since endogenously derived peptides (e.g. neoantigens) are presented to T lymphocytes through major histocompatibility complex (MHC) class I pathway,44 we next examined the RNA expression of HLA class I genes (i.e. HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G and B2M) in patients with high- versus low-ITH tumors. Intriguingly, the RNA expression of all HLA class I genes was significantly lower in patients with high- versus low-ITH tumors from patients with ccRCC from TCGA (HLA-A: p = 0.001; HLA-B: p = 0.006; HLA-C: p = 0.001; HLA-E: p = 0.002; HLA-F: p = 0.007; HLA-G: p = 0.024; B2M: p = 0.016; Figure 2E). Similar trends were also observed in ccRCC patients from Wang et al.,27 whether a MATH score of 41 (HLA-A: p = 0.015; HLA-B: p = 0.019; HLA-C: p = 0.020; HLA-E: p = 0.033; HLA-F: p = 0.022; HLA-G: p = 0.601; B2M: p = 0.035; Figure 2F) or 54 (HLA-A: p = 0.006; HLA-B: p = 0.024; HLA-C: p = 0.004; HLA-E: p = 0.002; HLA-F: p = 0.018; HLA-G: p = 0.269; B2M: p = 0.009; Figure 2G) was used to differentiate high- from low-ITH tumors, suggesting that immune escape might be more prevalent in high-ITH tumors from patients with ccRCC through HLA downregulation, an observation which had been described in lung squamous cell carcinoma.6
Given that tumor cells can evade immune surveillance by expressing programmed death ligand 1 (PD-L1) to bind programmed death-1 (PD-1) on the surface of activated T and B cells, thereby negatively regulating immune activity,45 we further evaluated the RNA expression of PD-L1 in high- versus low-ITH tumors. As a result, we found no significant difference in PD-L1 expression between tumors with low and high ITH (p = 0.066 for tumors from TCGA, p = 0.818 and 0.205 for those from Wang et al.;27 Figure S4A). Since PD-1 attenuates T-cell activation through interaction with PD-L1 and a second ligand—PD-L28, we also assessed the RNA expression of PD-L2 in high- versus low-ITH tumors. Similarly, no significant difference was observed in PD-L2 expression between tumors with low and high ITH (p = 0.657 for tumors from TCGA, p = 0.601 and 0.736 for those from Wang et al.;27 Figure S4B). Additionally, the RNA expression of PD-1 showed no significant difference in high- versus low-ITH tumors (Figure S4C). Overall, we found that low ITH was associated with more neoantigens, elevated expression of HLA class I genes, and higher abundance of DCs in ccRCC, suggesting increased immune activity in low-ITH tumors from patients with ccRCC.
Low ITH correlates with increased immune activity involving different immune compartments in pRCC
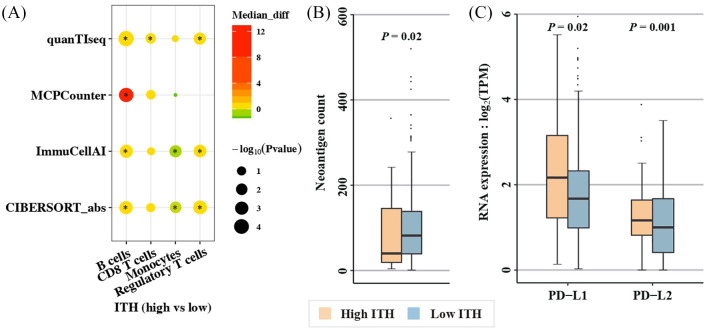
To determine whether ccRCC tumors share the above features with pRCC tumors, similar analysis was conducted in pRCC patients from TCGA. No significant difference was observed in DCs between high- and low-ITH tumors. However, low-ITH tumors from pRCC patients showed lower abundance of regulatory T cells (Tregs) and B cells (Tregs: p = 0.005 by quanTIseq, p = 0.0007 by CIBERSORT and p = 0.003 by ImmuCellAI; B cells: p = 0.0002 by MCPCounter, p = 6.8e-05 by quanTIseq, p = 0.001 by CIBERSORT and p = 0.001 by ImmuCellAI; Figure 3A), and trended to have lower abundance of CD8+ T cells (p = 0.03 by quanTIseq, p = 0.074 by MCPCounter and p = 0.084 by CIBERSORT; Figure 3A). Furthermore, low-ITH tumors displayed higher abundance of monocytes (p = 0.012 by CIBESORT and p = 0.005 by ImmuCellAI; Figure 3A) compared with high-ITH tumors.
Figure 3.
Low ITH correlates with increased immune activity involving different immune compartments in pRCC.
(A) Infiltration difference in high- versus low-ITH tumors (*denotes p < 0.05); (B) Neoantigen counts derived from both SNVs and InDels between tumors with high and low ITH; and (C) RNA expression of PD-1 ligands (PD-L1 and PD-L2) in high- versus low-ITH tumors. Tumors with MATH score ⩽31 were classified as low ITH, and high ITH otherwise.
ITH, intratumor heterogeneity; MATH, mutant-allele tumor heterogeneity; pRCC, papillary renal cell carcinoma; SNV, single nucleotide variation.
Apart from monocytes, neoantigen counts were also found to be significantly higher in low- versus high-ITH tumors (p = 9.7e-05 for neoAgs_SNVs, Figure S3A; p = 0.046 for neoAgs_InDels, Figure S3B; p = 0.02 for the combined neoantigen counts, Figure 3B). No significant association was observed between the level of ITH and the RNA expression of HLA class I genes (Figure S4D), but high-ITH tumors displayed significantly higher expressions of PD-L1 (p = 0.02; Figure 3C) and PD-L2 (p = 0.001; Figure 3C) compared with low-ITH tumors, indicating that immune escape might be more widespread in high-ITH tumors from patients with pRCC through upregulation of PD-1 ligands. Additionally, the RNA expression of PD-1 showed no significant difference in high- versus low-ITH tumors (Figure S3D). Together, these data suggest that low ITH is associated with increased immune activity in pRCC and that distinct mechanisms may be involved in tumors from different RCC subtypes to escape from immune surveillance.
Low ITH correlates with increased response to anti-PD-1 immunotherapy
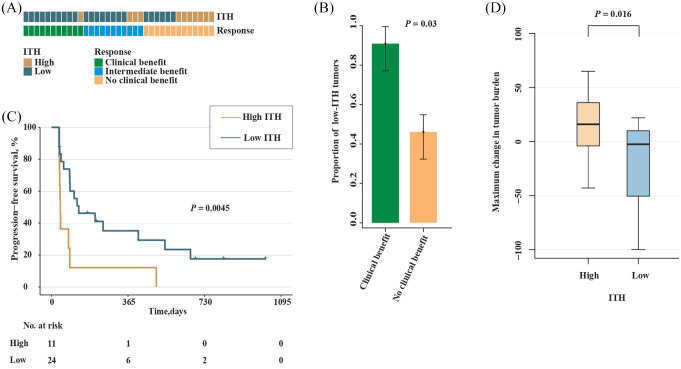
In light of the correlation between ITH and immune activity in ccRCC as well as pRCC tumors, we next examined whether there was a correlation between the level of ITH and patient survival after immunotherapy. We analyzed a data set derived from a recent clinical trial of anti-PD-1 therapy (nivolumab) in 35 patients with metastatic ccRCC.14 The number of patients in the clinical benefit (CB), intermediate benefit (IB) and no clinical benefit (NCB) groups were 11, 11 and 13, respectively (Table 1). We first assessed the level of ITH in patients within the CB or NCB group. As a result, low-ITH tumors were found to be enriched in patients from the CB versus NCB group (10/11 versus 6/13; p = 0.03; Fisher’s exact test; Figure 4A and B). Next, we examined survival in patients with low- versus high-ITH tumors. As expected, significantly prolonged PFS was observed in patients with low-ITH tumors compared with those with high-ITH tumors (HR, 0.32; 95% CI, 0.14–0.73; p = 0.0045; Figure 4C). We further compared the maximum change in tumor burden between patients with high- and low-ITH tumors and found that patients with low-ITH tumors experienced more reductions in tumor burden (p = 0.016; Figure 4D).
Figure 4.
Low ITH correlates with increased response to anti-PD-1 therapy in ccRCC.
(A) ITH and response to anti-PD-1 therapy by sample. (B) Proportion of low-ITH tumors in the CB versus NCB group. Error bars are standard errors, which were calculated as follows: , where p is the proportion in the population and n is the sample size. (C) PFS in high- versus low-ITH tumors from patients with metastatic ccRCC. (D) Maximum change in tumor burden between patients with high- and low-ITH tumors.
CB, clinical benefit; ccRCC, clear cell renal cell carcinoma; ITH, intratumor heterogeneity; NCB, no clinical benefit; PFS, progression-free survival.
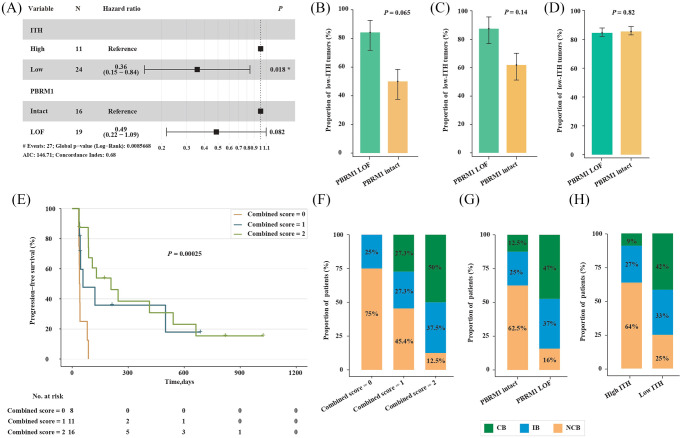
Since LOF mutations (i.e. frameshift insertion or deletion, nonsense mutation, and splice-site) in the PBRM1 gene were previously reported to correlate with response to ICT in the same data set,14 we next used a multivariable Cox proportional hazards model to include both the level of ITH and the mutation status of PBRM1 gene to determine whether low ITH was independent of PBRM1 LOF alterations in predicting better survival. In this multivariate model, the contribution of low ITH was significant (HR, 0.36; 95% CI, 0.15–0.84; p = 0.018; Figure 5A), demonstrating low ITH to be a predictor independent of PBRM1 LOF alterations. In addition, PBRM1 LOF alterations trended toward the prediction of better survival (HR, 0.49; p = 0.082; Figure 5A). There was no significant correlation between low ITH and PBRM1 LOF alterations, whether in the same data set (16/19 versus 8/16; p = 0.065; Fisher’s exact test; Figure 5B), or in patients with metastatic ccRCC from TCGA (14/16 versus 13/21; p = 0.14; Fisher’s exact test; Figure 5C) or in all ccRCC patients from TCGA (98/116 versus 188/220; p = 0.82; Chi-square test; Figure 5D).
Figure 5.
Association between the level of ITH and the mutation status of PBRM1 gene in ccRCC.
(A) Multivariate analysis of the level of ITH and the mutation status of PBRM1 gene. Proportion of low-ITH tumors in patients with versus without PBRM1 LOF alterations from the clinical trial of anti-PD-1 therapy (B) or TCGA (C and D). Error bars are standard errors, which were calculated as mentioned above. (E) PFS in patients with metastatic ccRCC who had different combined scores. The combined score was 2 if low ITH and PBRM1 LOF mutations simultaneously existed in a patient, 1 if either of them existed, and 0 if neither of them existed. The distribution of patients from the CB, IB and NCB group in patients with metastatic ccRCC who had different combined scores (F), or different mutation status of PBRM1 gene (G), or distinct level of ITH (H).
CB, clinical benefit; ccRCC, clear cell renal cell carcinoma; IB, intermediate benefit; ITH, intratumor heterogeneity; LOF, loss-of-function; NCB, no clinical benefit; PFS, progression-free survival; TCGA, The Cancer Genome Atlas.
To determine whether the combination of low ITH and PBRM1 LOF alterations could perform better than the individual predictors in predicting the benefit of anti-PD-1 immunotherapy in ccRCC patients, we next defined a combined score for each patient with metastatic ccRCC from the clinical trial of anti-PD-1 therapy.14 The combined score was 2 if low ITH and PBRM1 LOF mutations simultaneously existed in a patient, 1 if either one of them existed, and 0 if neither of them existed. As expected, higher combined scores correlated with better PFS (HR, 0.42; 95% CI, 0.24–0.73; p = 0.00025; Figure 5E). Notably, none (0/8) of the patients that had a combined score of zero was in the CB group, and only 12.5 percent (2/16) of the patients who had a combined score of two were in the NCB group (Figure 5F); whereas 12.5% (2/16) of the patients that did not have PBRM1 LOF alterations were in the CB group, and 16% (3/19) of the patients that had PBRM1 LOF alterations were in the NCB group (Figure 5G). In addition, 9% (1/11) of the patients with high-ITH tumors were in the CB group, and 25% (6/24) of the patients with low-ITH tumors were in the NCB group (Figure 5H). Together, these data suggest that the combination of the level of ITH and the mutation status of PBRM1 gene could perform better than the individual predictors in predicting the benefit of anti-PD-1 immunotherapy in patients with ccRCC.
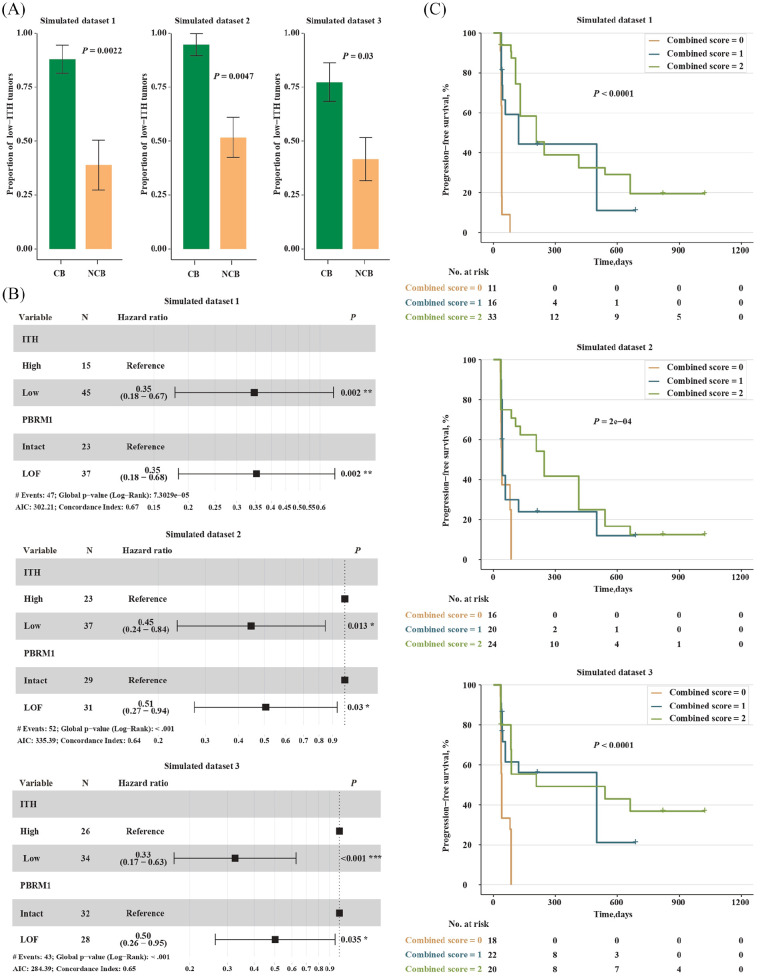
To validate the correlation between low ITH and the clinical benefit of PD-1 blockade in ccRCC, three equal-size simulated data sets (n = 60) were generated through random sampling with replacement based on the data from the clinical trial of anti-PD-1 in ccRCC.14 In the three data sets, the number of low-ITH tumors was 45, 37 and 34, respectively, and the number of patients who derived clinical benefit from the treatment of PD-1 blockade was 25, 19 and 22, respectively. Please see Table S2 for details. As expected, the enrichment of low-ITH tumors in the CB versus NCB group was observed in all the three data sets (22/25 versus 7/18, p = 0.0022; 18/19 versus 15/29, p = 0.0047; 17/22 versus 10/24, p = 0.03; Chi-square test; Figure 6A). Furthermore, low ITH was significantly associated with better PFS in all the three data sets, and the significance remained after including the mutation status of PBRM1 gene into a multivariable Cox proportional hazards model (Figure 6B). Possibly due to the larger sample size of the simulated data sets, the contribution of PBRM1 LOF alterations to better survival was significant in the multivariable model (Figure 6B). In addition, patients with higher combined scores exhibited prolonged PFS in all the three data sets (Figure 6C). Overall, these data indicate that the level of ITH could be used to predict benefit of anti-PD-1 immunotherapy in RCC.
Figure 6.
Validation in simulated data sets.
(A) Proportion of low-ITH tumors in the CB versus NCB group. Error bars are standard errors, which were calculated as mentioned above; (B) Multivariate analysis of the level of ITH and the mutation status of PBRM1 gene; and (C) PFS in patients with metastatic ccRCC who had different combined scores.
CB, clinical benefit; ccRCC, clear cell renal cell carcinoma; ITH, intratumor heterogeneity; NCB, no clinical benefit; PFS, progression-free survival.
Discussion
A key unanswered question in RCC is whether and which genomic metrics are correlated with clinical benefit of ICT. Our results suggest that low ITH is associated with increased response to anti-PD-1 immunotherapy in RCC through increased immune activity involving more neoantigens and less frequent immune evasion.
We uncovered a negative relationship between ITH and neoantigen count in ccRCC tumors. This may seem to be counterintuitive since tumors with high ITH are generally assumed to have more somatic mutations and thus more neoantigens. Nevertheless, direct evidence for the restriction of ITH by adaptive immune response has been recently provided by Milo et al. in lymphoma.46 In line with these observations, we also found increased expression of HLA class I genes and higher abundance of DCs in low-ITH tumors from patients with ccRCC. Intriguingly, MHC class I peptide complexes binding to T-cell receptors with sufficient affinity are always presented to T cells by DCs.47 That is to say, tumors with high ITH may have more neoantigens, yet the high neoantigen load may in turn work in concert with immune system to change the level of ITH in these tumors from high to low.
We also revealed an inverse association between ITH and immune infiltration in ccRCC tumors, which was in agreement with previous studies.5,21–23 Among the immune cells, DCs showed the same trend in ccRCC tumors from TCGA and Wang et al.27 The same is true of NK cells when a MATH score of 54 was used to perform ITH classification for ccRCC tumors from Wang et al.27 In addition, ccRCC tumors from TCGA exhibited inverse associations of ITH with monocytes and neutrophils, while those from Wang et al.27 displayed negative relationship between ITH and B cells. Notably, NK cells are innate immune effectors which spontaneously kill cells considered to be dangerous to the host (e.g. cancer cells).48 Furthermore, two different studies recently revealed the association of DCs with response to ICT,39,41 one of which included RCC patients.41 Taken together, the data mentioned above point to a potential relationship between ITH and response to ICT in patients with ccRCC.
Using published data from a clinical trial of anti-PD-1 immunotherapy in patients with metastatic ccRCC, we found low ITH to be associated with increased response to PD-1 blockade, which was consistent with previous findings in other cancer types.7,40 Noteworthily, the association was independent of PBRM1 LOF alterations, a recently identified marker of response to ICT in ccRCC.14,15 Possibly due to the small sample size of the data set, the contribution of PBRM1 LOF alterations in the multivariable analysis was not statistically significant although PBRM1 LOF alterations trended toward better survival. Therefore, we expanded the sample size from 35 to 60 when generating the simulated data sets, and better survival was significantly associated with both low ITH and PBRM1 LOF alterations in the multivariable analysis. More significantly, whether in the clinical trial of anti-PD-1 immunotherapy or the simulated data sets, the combination of the level of ITH and the mutation status of PBRM1 gene performed better than the individual predictors in predicting benefit of anti-PD-1 immunotherapy in patients with ccRCC.
In pRCC, we also revealed a negative relationship between ITH and neoantigen count. However, decreased expression of PD-1 ligands (i.e. PD-L1 and PD-L2), instead of increased expression of HLA class I genes, was observed in low- versus high-ITH tumors from pRCC, suggesting more frequent immune evasion in high-ITH tumors and distinct mechanisms to evade immune surveillance between high-ITH tumors from pRCC (upregulation of PD-1 ligands) and ccRCC (downregulation of HLA class I genes). Notably, positivity of either PD-L1 or PD-L2 has been previously reported to be associated with shorter survival,49,50 indicating the association between low ITH and improved outcome in pRCC. Meanwhile, several types of immune cells were differentially distributed between tumors with high and low ITH, with ITH displaying a negative relationship with monocytes, yet positive associations with B cells, CD8+ T cells and Tregs. As for tumor-infiltrating B cells in RCC, some studies suggest that they are metastasis-promoting51 and associated with poor prognosis,52 whereas a positive association with better response to targeted therapy has also been shown.53 The same is true for monocytes, different subsets of which perform functions that result in both pro- and anti-tumoral immunity.54 Nevertheless, tumor tissues infiltrating large amounts of Tregs are usually associated with poor prognosis,55 and an increased level of CD8+ T cells have also been reported to be associated with worse outcome in RCC,56 further supporting improved outcome in low- versus high-ITH tumors in pRCC. Interestingly, monocytes, as precursors of DCs,54 have been recently shown to be associated with clinical benefit of anti-PD-1 immunotherapy.57 Given the equally increased immune activity (i.e. more neoantigens and less frequent immune evasion) in low-ITH tumors from pRCC patients, the level of ITH may also be able to predict survival after anti-PD-1 immunotherapy in pRCC where no genomic metrics has been found to correlate with response to ICT, although further data are needed to confirm these observations.
In this study, we quantified ITH using MATH16 based on the allele frequencies of somatic mutations. Admittedly, there are other well-designed tools (e.g. PyClone,2 EXPANDS3 and ABSOLUTE1) quantifying ITH based on not only somatic mutations, but also other metrics such as copy number estimates and loss of heterozygosity. While taking more parameters into account may improve the accuracy of ITH quantification, it also means more complex analyses and longer run time, which is not conducive to large-scale clinical application. Furthermore, the association of high ITH with poor outcome revealed by EXPANDS4 or PyClone,5 as well as the inverse association between ITH and immune cell infiltration revealed by PyClone,5 has also been identified using MATH in this study and previous studies.17–22 Additionally, MATH score was positively correlated with subclonal genome fraction determined by ABSOLUTE1 in this study. Therefore, MATH could be a fast and accurate measurement of ITH in RCC.
Notably, the cutpoint of MATH scores to distinguish high- from low-ITH tumors was 31 and 41 for pRCC and ccRCC patients from TCGA, respectively, and 42 for patients with metastatic ccRCC from a clinical trial of anti-PD-1 therapy. These data suggest that the cutpoint of MATH scores varies with tumor types/subtypes. Aligning with this observation, the cutpoint of MATH scores was previously reported to be 32 for head and neck cancer,17 and 23.9 for early stage diffuse large B-cell lymphoma.20 As for large-scale clinical application, the cutpoint of MATH scores for ccRCC and pRCC need further preclinical and prospective clinical validation.
There are several shortcomings of our study. Firstly, the exact cutpoint of MATH scores was not determined for ccRCC tumors from Wang et al.27 due to the lack of survival information. To address this issue, both the cutpoint determined for ccRCC tumors from TCGA and the MATH value that best differentiate high from low subclonal genome fraction in tumors from Wang et al.27 were applied to perform ITH classification for this data set, and major findings observed in ccRCC tumors from TCGA were also observed in those from Wang et al.27 Secondly, the RNA-seq data were only available in about two-thirds (98/152) of the ccRCC tumors from Wang et al.27 To address this issue, we employed four different tools for immune deconvolution, and a type of immune cell would be considered differentially distributed only if the same trend was observed in the results of at least two tools. Thirdly, another whole-exome sequencing data set for ccRCC patients who were treated with PD-1 blockade was not available to further confirm the association of ITH with clinical benefit of PD-1 inhibitors in ccRCC. To address this issue, we generated three equal-size simulated data sets based on the clinical trial of anti-PD-1 therapy in patients with metastatic ccRCC,14 and finally validated the association between low ITH and response to PD-1 blockade in ccRCC. In addition, as a relatively rare subtype of RCC, an independent validation cohort was not available for pRCC to confirm our findings in pRCC patients from TCGA. We hope that more pRCC samples treated with or without PD-1 blockade will be sequenced and made publicly available to confirm the association of ITH with immune activity and response to PD-1 inhibitors.
In summary, our data suggest that assays of ITH may be useful for determining which RCC patients are most likely to respond to PD-1 blockade. Furthermore, understanding the mechanism by which ITH affects response to PD-1 inhibitors may provide insights into how ITH might affect cancer therapy and patient survival, and help to develop predictive biomarkers for response to ICT.
Supplemental Material
Supplemental material, sj-pdf-1-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-3-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology
Acknowledgments
We thank Eliezer M. Van Allen for patiently explaining the details of the statistical analyses in his recently published study and for helpful advice.
Footnotes
Authors’ contributions: Conception and design: XR and ZS. Development of methodology: XR. Acquisition of data: XR. Analysis and interpretation of data (e.g. statistical analysis, biostatistics, computational analysis): XR, JX, YZ, and FC. Writing, review, and/or revision of the manuscript: XR, HT, HC, and ZS. Administrative, technical, or material support (i.e. reporting or organizing data, constructing databases): XR, JX, YZ, HT, KZ, and ZS. Study supervision: XR and ZS. All authors read and approved the final manuscript.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Key Research and Development Program of China (2016YFC0900400).
Conflict of interest statement: The authors declare that there is no conflict of interest.
Data availability: Data supporting the results reported in the article have been provided in supplementary materials/tables.
Supplemental material: Supplemental material for this article is available online.
ORCID iD: Zhongsheng Sun  https://orcid.org/0000-0003-1992-2681
https://orcid.org/0000-0003-1992-2681
Contributor Information
Xia Ran, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China; Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing, China; CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing, China.
Jinyuan Xiao, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China.
Yi Zhang, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China.
Huajing Teng, Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing, China.
Fang Cheng, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China.
Huiqian Chen, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China.
Kaifan Zhang, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, China.
Zhongsheng Sun, Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou, Zhejiang 325000, China; Beijing Institutes of Life Science, Chinese Academy of Sciences, Beichen West Road, Chao Yang District, Beijing, 100101, China.
References
- 1. Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014; 11: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andor N, Harness JV, Muller S, et al. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics 2014; 30: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016; 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morris LG, Riaz N, Desrichard A, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 2016; 7: 10051–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McGranahan N, Furness AJS, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016; 351: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolf Y, Bartok O, Patkar S, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 2019; 179: 219–235.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Turajlic S, Xu H, Litchfield K, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 2018; 173: 595–610.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haddad AQ, Margulis V. Tumour and patient factors in renal cell carcinoma-towards personalized therapy. Nat Rev Urol 2015; 12: 253–262. [DOI] [PubMed] [Google Scholar]
- 10. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin 2017; 67: 507–524. [DOI] [PubMed] [Google Scholar]
- 11. Cancer Genome Atlas Research Network, Linehan WM, Spellman PT, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016; 374: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017; 377: 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Legrand FA, Gandara DR, Mariathasan S, et al. Association of high tissue TMB and atezolizumab efficacy across multiple tumor types. J Clin Oncol 2018; 36: 12000. [Google Scholar]
- 14. Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018; 359: 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Braun DA, Ishii Y, Walsh AM, et al. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol 2019; 5: 1631–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 2013; 49: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mroz EA, Tward AD, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 2015; 12: e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang F, Wang Y, Li Q, et al. Intratumor heterogeneity predicts metastasis of triple-negative breast cancer. Carcinogenesis 2017; 38: 900–909. [DOI] [PubMed] [Google Scholar]
- 19. Zhang JY, Yan SC, Liu XY, et al. Gender-related prognostic value and genomic pattern of intra-tumor heterogeneity in colorectal cancer. Carcinogenesis 2017; 38: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Feng W, Liu P. Genomic pattern of intratumor heterogeneity predicts the risk of progression in early stage diffuse large B-cell lymphoma. Carcinogenesis 2019; 40: 1427–1434. [DOI] [PubMed] [Google Scholar]
- 21. Karn T, Jiang TT, Hatzis C, et al. Association between genomic metrics and immune infiltration in triple-negative breast cancer. JAMA Oncol 2017; 3: 1707–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Safonov A, Jiang T, Bianchini G, et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Res 2017; 77: 3317–3324. [DOI] [PubMed] [Google Scholar]
- 23. McDonald KA, Kawaguchi T, Qi Q, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol 2019; 26: 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019; 35: i436–i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pachter L. Models for transcript quantification from RNA-Seq. arXiv preprint arXiv:1104.3889. [Google Scholar]
- 26. Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015; 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang XM, Lu Y, Song YM, et al. Integrative genomic study of Chinese clear cell renal cell carcinoma reveals features associated with thrombus. Nat Commun 2020; 11: 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 2011; 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018; 15: 591–594. [DOI] [PubMed] [Google Scholar]
- 31. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018; 48: 812–830.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016; 12: e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schenck RO, Lakatos E, Gatenbee C, et al. NeoPredPipe: high-throughput neoantigen prediction and recognition potential pipeline. BMC Bioinformatics 2019; 20: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szolek A, Schubert B, Mohr C, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014; 30: 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016; 17: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finotello F, Mayer C, Plattner C, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019; 11: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miao Y-R, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020; 7: 1902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018; 50: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mayoux M, Roller A, Pulko V, et al. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci Transl Med 2020; 12: eaav7431. [DOI] [PubMed] [Google Scholar]
- 42. Matsumoto K, Fukuda N, Abe M, et al. Dendritic cells and macrophages in kidney disease. Clin Exp Nephrol 2010; 14: 1–11. [DOI] [PubMed] [Google Scholar]
- 43. Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017; 18: 1009–1021. [DOI] [PubMed] [Google Scholar]
- 44. Jongsma MLM, Guarda G, Spaapen RM.1. The regulatory network behind MHC class I expression. Mol Immunol. Epub ahead of print 8 December 2017. DOI: 10.1016/j.molimm.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 45. Iwai Y, Ishida M, Tanaka Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A 2002; 99: 12293–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Milo I, Bedora-Faure M, Garcia Z, et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci Immunol 2018; 3: eaat1435. [DOI] [PubMed] [Google Scholar]
- 47. Yewdell JW, Dersh D, Fahraeus R. Peptide channeling: the key to MHC class I immunosurveillance? Trends Cell Biol 2019; 29: 929–939. [DOI] [PubMed] [Google Scholar]
- 48. Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol 2016; 17: 1025–1036. [DOI] [PubMed] [Google Scholar]
- 49. de Velasco G, Miao D, Voss MH, et al. Tumor mutational load and immune parameters across metastatic renal cell carcinoma risk groups. Cancer Immunol Res 2016; 4: 820–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shin SJ, Jeon YK, Kim PJ, et al. Clinicopathologic analysis of PD-L1 and PD-L2 expression in renal cell carcinoma: association with oncogenic proteins status. Ann Surg Oncol 2016; 23: 694–702. [DOI] [PubMed] [Google Scholar]
- 51. Li S, Huang C, Hu G, et al. Tumor-educated B cells promote renal cancer metastasis via inducing the IL-1β/HIF-2α/Notch1 signals. Cell Death Dis 2020; 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sjoberg E, Frödin M, Lövrot J, et al. A minority-group of renal cell cancer patients with high infiltration of CD20+B-cells is associated with poor prognosis. Br J Cancer 2018; 119: 840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin Z, Liu L, Xia Y, et al. Tumor infiltrating CD19+ B lymphocytes predict prognostic and therapeutic benefits in metastatic renal cell carcinoma patients treated with tyrosine kinase inhibitors. Oncoimmunology 2018; 7: e1477461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. J Leukoc Biol 2019; 106: 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res 2017; 27: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drake CG, Stein MN. The immunobiology of kidney cancer. J Clin Oncol. Epub ahead of print 29 October 2018. DOI: 10.1200/JCO.2018.79.2648. [DOI] [PubMed] [Google Scholar]
- 57. Krieg C, Nowicka M, Guglietta S, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 2018; 24: 144–153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-2-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology
Supplemental material, sj-xlsx-3-tam-10.1177_1758835920977117 for Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma by Xia Ran, Jinyuan Xiao, Yi Zhang, Huajing Teng, Fang Cheng, Huiqian Chen, Kaifan Zhang and Zhongsheng Sun in Therapeutic Advances in Medical Oncology