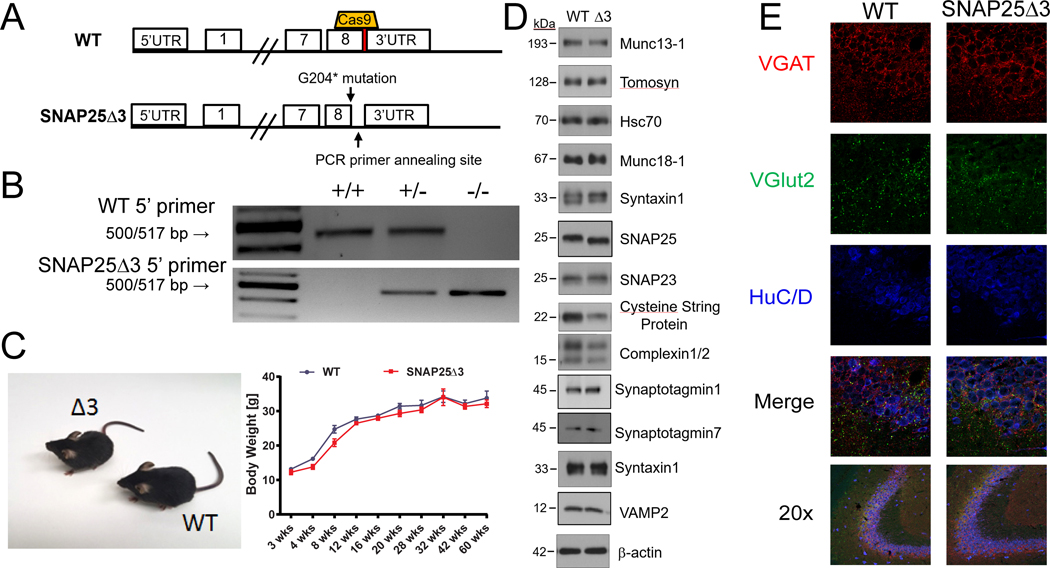
Fig. 1. Generation of the SNAP25Δ3 mouse by CRISPR-Cas9.
(A) Graphical representation of the region on mouse chromosome 2 targeted by the sgRNA (top) cloned into px330 and the subsequent region after homology-directed repair (bottom) containing the G204* mutation and cloning site. (B) Agarose gel electrophoresis of PCR products generated from reactions containing two different 5’ primers used to genotype WT, heterozygote, and homozygote SNAP25Δ3 littermate animals. The WT 5’ primer corresponds to the WT region on mouse chromosome 2, whereas the SNAP25Δ3 primer corresponds to the region containing the G204* mutation. (C) Gross morphology of WT and SNAP25Δ3 homozygotes (left) and growth curves (right) showing the increase in body mass in WT mice (n = 12 to 18 mice) and SNAP25Δ3 homozygotes (n = 12 to 21 mice) over the first 60 weeks of life of several noncontinuous cohorts. (D) Western blotting analysis of presynaptic proteins found within synaptosomal (93,99, 100) fractions (all except for syt I and VII, and VAMP2) or whole mouse brain lysate (syt I and VII, and VAMP2) of WT and SNAP25Δ3 (Δ3) mice. n= 3 to 12 biological replicates per condition. The abundance of cysteine string protein (CSP) in the presynaptic fraction of SNAP25Δ3 synaptosomes was reduced compared to that in WT synaptosomes (see Supplemental Figure 1). No statistically significant difference was found in any of the other proteins. (E) Immunofluorescence imaging of GABAergic (VGAT) and glutamatergic (vGlut2) immunoreactive appositions(46) within hippocampal slices taken from adult WT or SNAP25Δ3 homozygotes. Data are representative of four mice per genotype. HuC/D was used as a pan-neuronal marker.