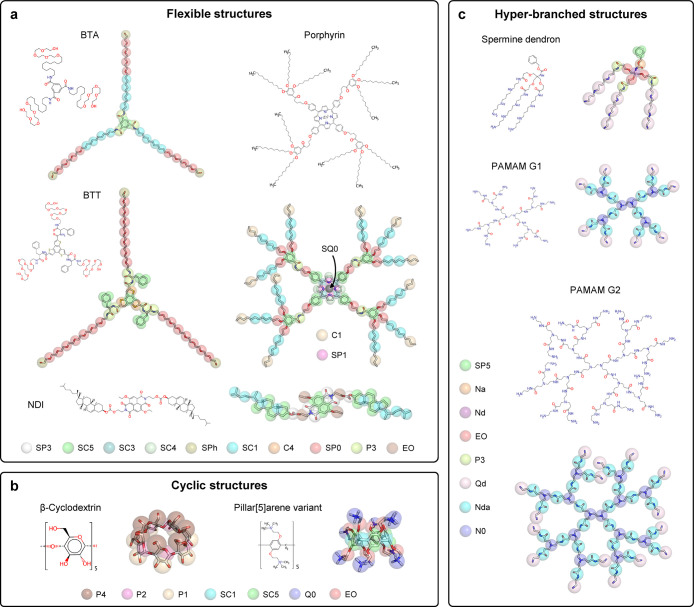
Figure 1.
Molecules used to benchmark Swarm-CG. Each molecule is represented by its molecular structure and AA model with superimposed CG MARTINI bead mapping. (a) Flexible and symmetric molecular structures generating supramolecular polymers: water-soluble BTA with amphiphilic side chains,83 C3-symmetric BTT decorated by l-phenylalanine and octaethylene glycol side-chains,84 NDI-based,47 and Zn-porphyrin-based molecules.50 (b) Examples of cyclic structures: β-cyclodextrin97 and a pillar[5]arene.98 (c) Complex hyper-branched polymer structures: spermine dendron99 and PAMAM G1 and G2.49,100−102 Each panel indicates the color coding of the CG MARTINI bead types (see Supporting Information for exact mapping data).