Figure 2. Eye phenotype of pax6 crispants.
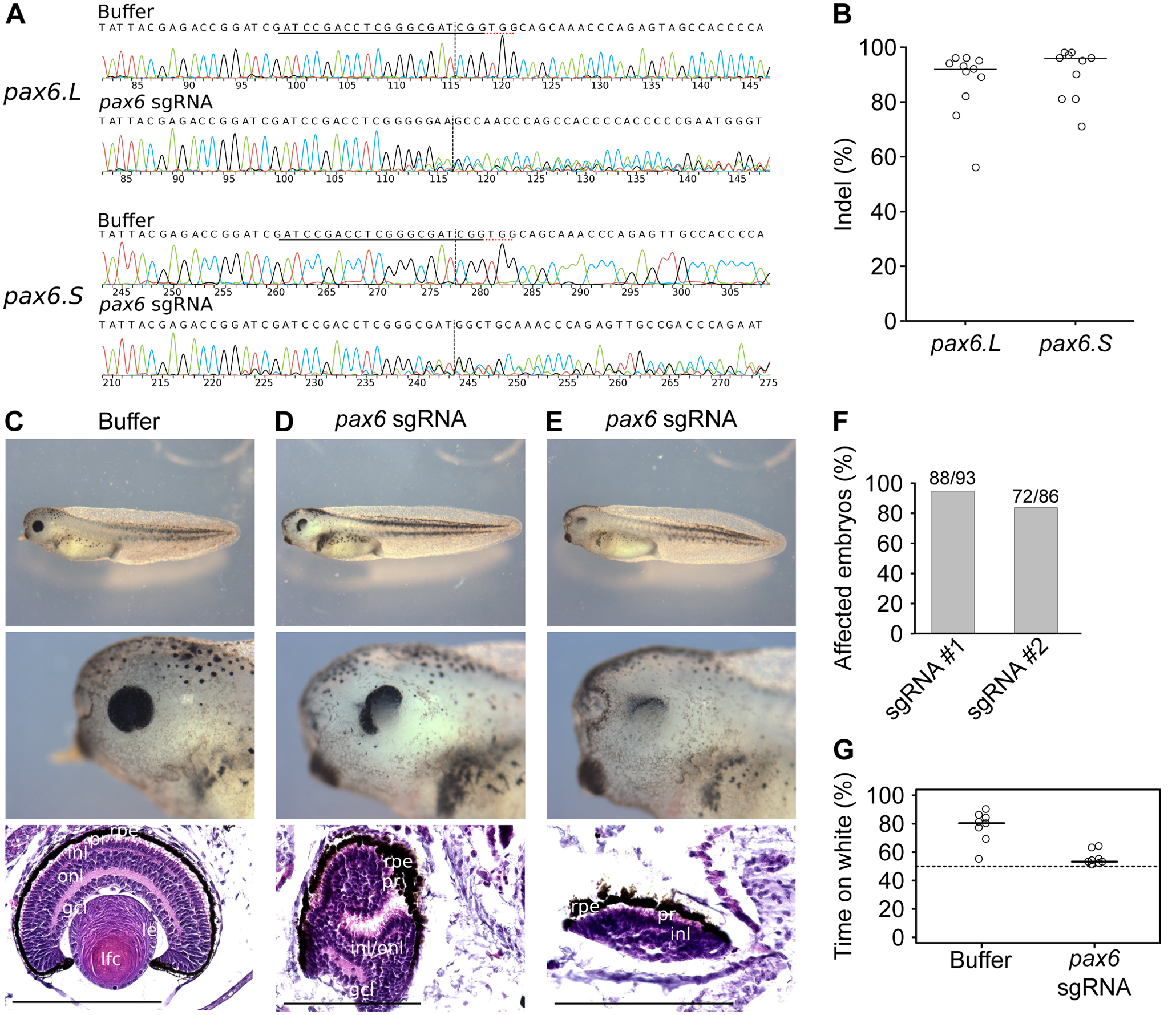
We injected embryos with Cas9 enzyme and a sgRNA targeted against pax6.L and pax6.S or buffer, and we allowed the embryos to develop until stage 41–42. A, We separately amplified the pax6.L and pax6.S loci from total embryos for Sanger sequencing. We show representative chromatograms of one sgRNA and one buffer-injected embryo. The sgRNA and PAM sequences are underlined and the cleavage sites are shown. B, The percentages of InDels in individual embryos were calculated from Sanger chromatograms with ICE37. C-E, from top to bottom whole embryo, higher magnification of the eye and histological section of the eye, scale bar 200 μm. C, Buffer-injected embryo. D-E, Two Cas9 and sgRNA injected embryos. Key for lens and retina layers, see Figure 1. F, Quantification of eye defects, in tadpoles previously injected with the same sgRNA against pax6 as above (sgRNA #1) or an alternative sgRNA against pax6 (sgRNA #2) G, Functional vision assay of stage 45 buffer-injected embryos and pax6 crispants (sgRNA #1). We put 10 tadpoles in a water tank with a black and a white side, we switched the sides and we counted the number of embryos on the white side after one minute. We repeated the switching-counting procedure 10 times for each batch of 10 tadpoles. The cumulative number of embryos on white (between 0 and 100) is shown here, for 8 independent pools of 10 tadpoles. The tadpoles spend more time on white if they are able to distinguish white from black, and the bias in favour of the white side reflects their visual acuity.
