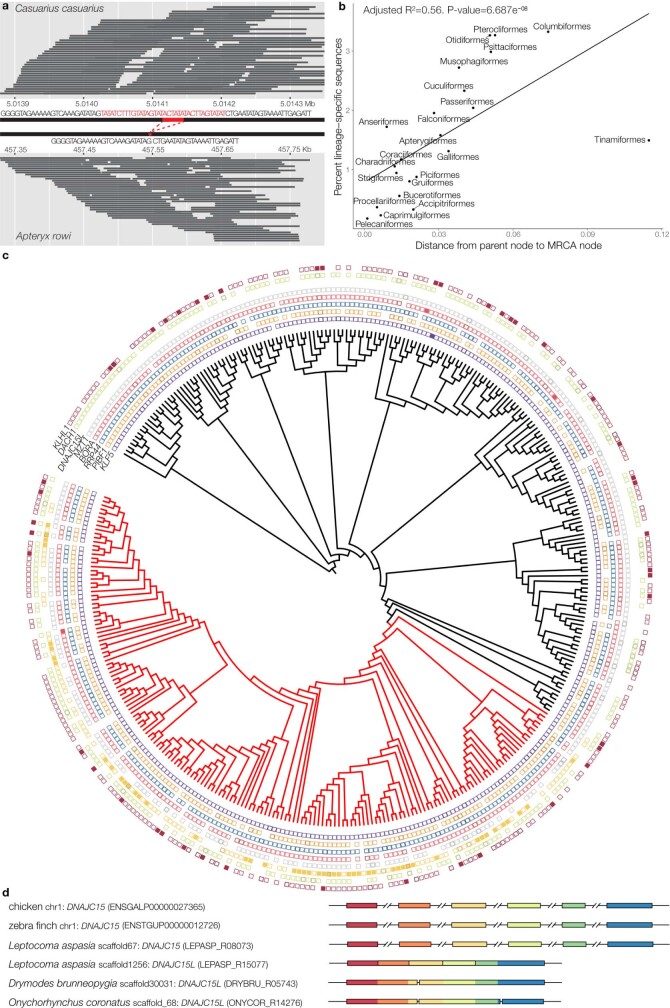
Extended Data Fig. 7. Identification of lineage-specific sequences.
a, An example of a 36-bp insertion (red) identified by Cactus in the southern cassowary (Casuarius casuarius) compared to the Okarito brown kiwi (Apteryx rowi) (both in Palaeognathae) with mapped sequence reads shown as lines. b, Proportion of lineage-specific sequence for each order correlated with the distance from parent node to MRCA node (branch length). c, Presence and absence of the DNAJC15-like gene (DNAJC15L), and its surrounding genes, in all 363 birds. Upstream: KLHL1 and DACH1; downstream: MZT1, BORA, RRP44, PIBF1 and KLF5. The state is shown for each bird in three ways: multiple copies (filled shapes), one copy (empty shapes) and no gene (blank). Passeriformes are highlighted in red. A zoomable figure with labels for all terminals is available at www.doi.org/10.17632/fnpwzj37gw. d, Exon fusion patterns of the DNAJC15-like gene (DNAJC15L) in three Passeriformes, compared to exon structure of the ancestral DNAJC15. For L. aspasia, gene models for the ancestral and novel copy are shown. The structure of the ancestral copy is highly conserved across all bird species with five introns. The Passeriformes-specific copy has no intron or newly derived minor intron and includes a poly-(A) at the 5′ end, which implies that this new gene was derived from retroduplication of DNAJC15.