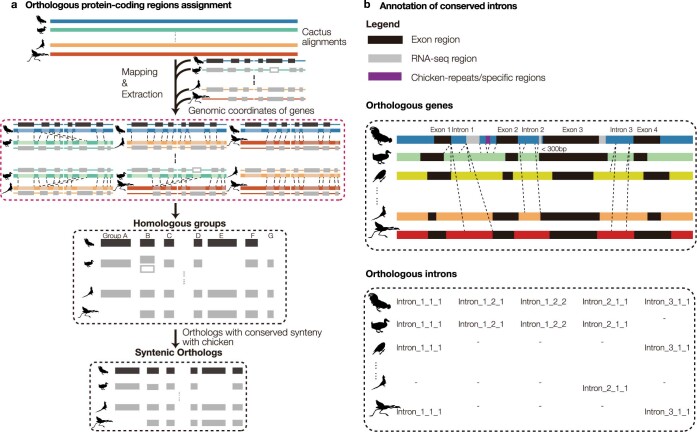
Extended Data Fig. 5. Overview of the pipelines for identifying genomic regions.
a, Assignment of orthologous protein-coding regions. All pairwise relationships between homologous regions obtained from the Cactus alignment (4 species shown here in different colours) were used to construct the homologous groups across all 363 birds. Using chicken as the reference, we further generated a table containing homologues with conserved synteny to chicken. b, Annotation of conserved orthologous intron regions on the basis of Cactus whole-genome alignments. The credible intron fragments in chicken were picked out after filtering out regions mapped by RNA sequences, and chicken-specific or repetitive regions. Orthologous relationships of intron fragments were detected on the basis of the aligned Cactus hits and the orthologues with conserved synteny with chicken. The non-intron regions of each bird in the alignments were masked as gaps.