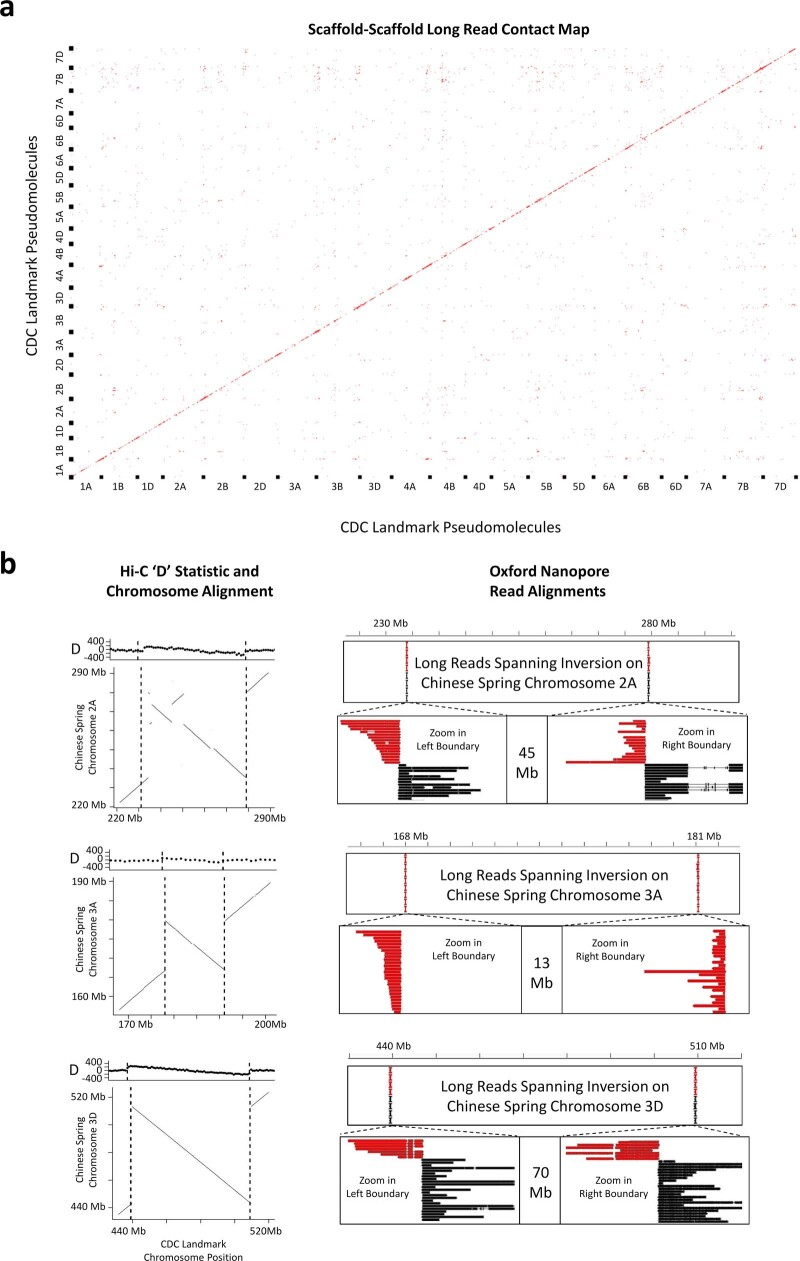
Extended Data Fig. 2. Evaluation of the CDC Landmark RQA using Oxford Nanopore Long Reads.
a, Scaffold-scaffold long read contact map showing shared read IDs between scaffold ends along the ordered scaffolds in the CDC Landmark pseudomolecules. The diagonal pattern indicates that adjacent scaffolds share the same long reads and are therefore properly ordered and oriented by Hi-C in the RQA. b, Characterization of inversion events on chromosomes 2A, 3A, and 3D. The directionality biases estimated from alignments of Hi-C data against Chinese Spring (left, top), and chromosome alignment of the inversion events between CDC Landmark and Chinese Spring RQAs (left, bottom) are shown. Long reads spanning the inversion events and magnified views of the reads aligning to the left and right boundaries of the inversions (right) are provided.