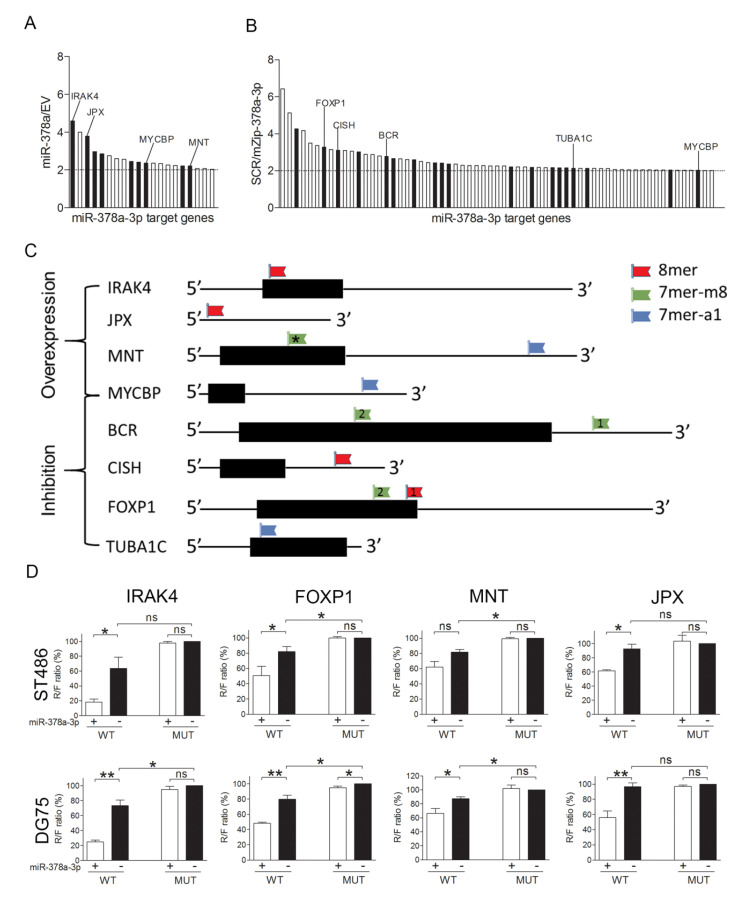
Figure 3.
Identification and validation of miR-378a-3p targets. MiR-378a-3p targets identified by Ago2-RIP-Chip upon (A) miR-378a overexpression relative to empty vector (EV) and (B) scrambled vector relative to miR-378a-3p inhibition (SCR/mZip-378a). The black bars indicate genes with miR-378a-3p seed binding sites. (C) Schematic representations of the eight genes selected for luciferase reporter assay validation. Black boxes indicate positions of the open reading frames (ORFs). Positions and types of miR-378a-3p binding sites are indicated relative to the ORF. The binding site in MNT indicated by an asterisk was not tested. (D) Luciferase reporter assay results upon co-transfection of ST486 and DG75 cells with the Psi-check-2 construct containing the wildtype (WT) or mutated (MUT) miR-378a-3p binding sites from the selected genes and either an miR-378a-3p mimic or a negative control mimic. Significant differences were calculated using a paired t-test. * p < 0.05, ** p < 0.01, ns = not significant.