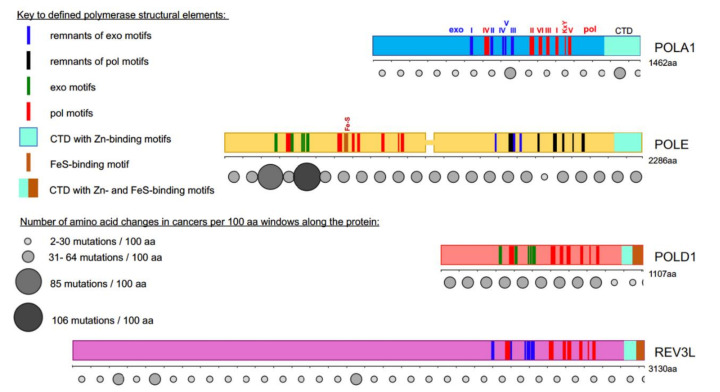
Figure 1.
Most cancer-associated mutations affect the catalytically active half of POLE. Colored bars represent the main subunits of DNA pols, a catalytic subunit of pol α, POLA1, in light blue; of pol ε, POLE, in yellow; of pol δ, POLD1, in red; and pol ζ, REV3L, in purple. Note that POLE is a tandem of active pol (N-terminal half) and inactive pol (C-terminal half) [5,6]. Evolutionarily conserved motifs characteristic for all exonuclease (exo) domains, are labeled I-V in green and pol domains are labeled I-VI and KxY in red [5,6,7,8,9,10,11,12]. The order of the motifs along all four proteins is the same, but they occupy different parts of the whole protein. For example, REV3L has a very long N-terminal part not related to pols. In POL1, REV3L, and the C-terminal half of POLE, the exonuclease motifs are inactivated during evolution; they are shown in blue. Inactivated pol motifs in the C-terminal half of POLE are shown in black. The key for these and other elements of the pol primary structure is in the left upper quarter of the figure. Rows of circles of different sizes and shades of grey below the catalytic pol subunits represent the number of missense mutations found in tumors along the protein regions in 100 amino acid increments. Variants were collected from the cBioPortal database from a curated non-overlapping collection of tumor genomes (https://www.cbioportal.org/ (cbioportal.org)). A guide explaining the relation between size and intensity of grey to the number of mutations found in the database in the 100 amino acids interval is on the left lower quarter of the figure.