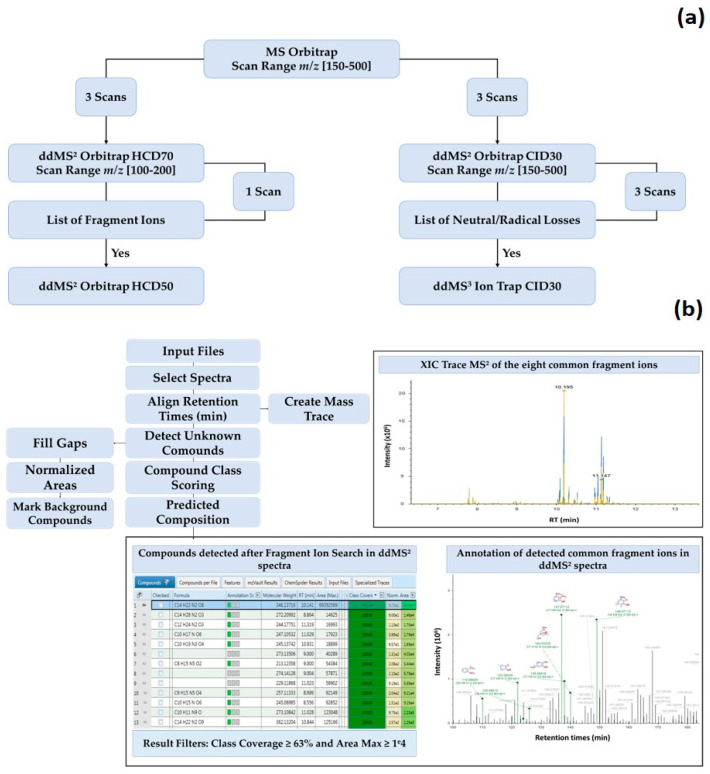
Figure 1.
Flowchart of the developed untargeted screening approach of MAAs using an Orbitrap MS based on data-dependent MS2/MS3 Acquisition (a). Untargeted workflow designed on Compound DiscovererTM to flag putative MAAs in algal extracts (b). Description of the role of the following nodes: Compound Class Scoring: Calculation of the percentage of common fragment ions detected in mass spectral data of every compound in the result table. Predicted Composition: Proposition of chemical formula for unknown compounds. Create mass Trace: Create mass chromatogram of compounds for which common fragment ions were detected in their mass spectral data. Fill Gaps: Indication of missing peaks or peaks below the detection threshold. Normalize Areas: Normalization of chromatographic peaks. Mark Background Compounds: Identification of compounds in blanks.