Figure 3.
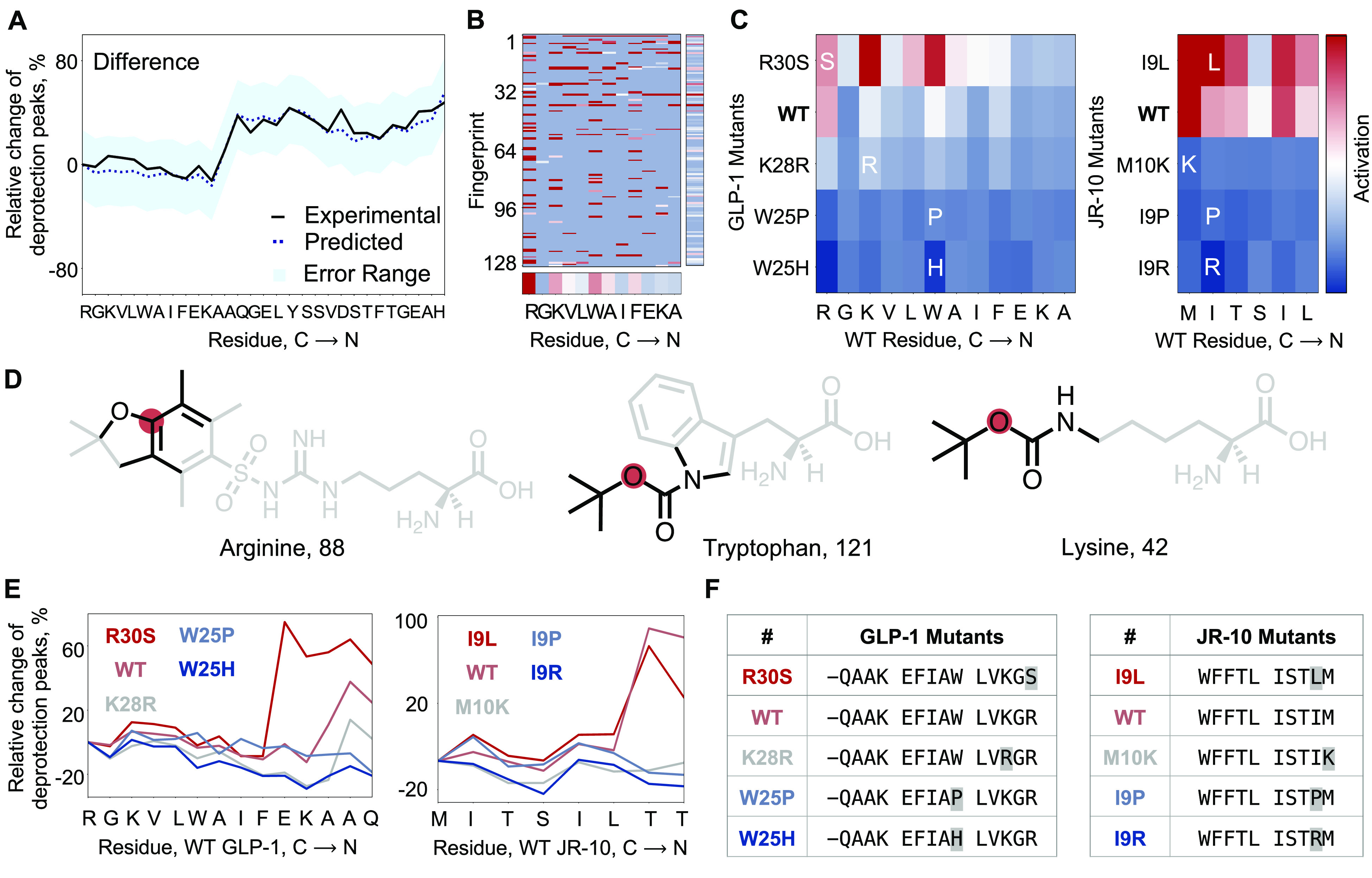
The deep-learning model predicts, interprets, and optimizes aggregation. (A) Predicted difference (width – height) is overlaid on the calculated difference from the experimentally obtained UV–vis deprotection trace for GLP-1. The predicted difference is within the error for the experimentally observed difference. Aggregation is defined as the step where the difference between width and height is greater than 20%. (B) Positive activation gradient map for GLP-1 prechain prior to the addition of third Ala (A18). The mean activation values for individual amino acids and bit-vectors are shown along respective axes. (C) Positive activation gradient maps averaged over fingerprint indices for GLP-1 and JR-10 mutants show a sharp decrease in aggregation from the negative control (GLP-1, R30S; JR-10, I9L) to the wild-type and the other mutants. The prechains considered in the analysis are for the known aggregating regions in GLP-1 (addition of third Ala, A18) and JR-10 (addition of second Thr, T4). The most activated amino acids are Arg, Trp, and Lys in WT GLP-1, and Met and Ile in WT JR-10. (D) Most activated substructures by amino acid for GLP-1 are shown. Amino acids with aryl groups and bulkier side-chain protecting groups are found to be most activated. The analysis excluded substructures in the amino acid scaffold, both the amide backbone and the side chains native to the respective amino acid. The red dot is the node atom, and the black bonds/atoms represent the chemical substructure encoded in the activated fingerprint. (E) Calculated difference from the experimental synthesis run for predicted sequence analogues of WT GLP-1 and WT JR-10. The analogues are predicted single-point mutations of the sequence—K28R, W25P, and W25H for GLP-1, and M10K, I9P, and I9R for JR-10. The predicted negative controls are R30S for GLP-1 and I9L for JR-10. The predicted sequence analogues, except negative controls, are less aggregating at the respective step. Negative control for GLP-1 is more aggregating than GLP-1 itself. Negative control for JR-10 is less aggregating than JR-10, but more aggregating than the other analogues. (F) Predicted GLP-1 and JR-10 mutants which were experimentally validated are listed. All mutants predicted using the model contain the mutation before the aggregating step, i.e., addition of third Ala for GLP-1, and addition of second Thr for JR-10. The in silico generation of mutants had no such constraints.
