Abstract
Sequence type 59 (ST59) is the dominant type of community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Taiwan. Previously, we reported that ST59 MRSA harbors enterococcal IS1216V-mediated multidrug-resistant composite transposons MESPM1 or MES6272-2. The MES were found to have a mosaic structure, largely originating in enterococci and partly native to S. aureus. The current study aimed to track the origin of the MES and how they disseminated from enterococci to ST59 S. aureus. A total of 270 enterococcal isolates were analyzed, showing that two ST64 Enterococcus faecalis isolated in 1992 and 11 clonal complex 17 Enterococcus faecium harbored MESPM1-like and MES6272-2-like structures, respectively. Sequence analysis revealed that ST64 E. faecalis strain N48 acquired the MESPM1-like structure on the plasmid pEflis48. The pEflis48 harbored the enterococci-originated region (erythromycin, kanamycin, and streptomycin resistances) and the S. aureus-originated region (chloramphenicol resistance) of MESPM1 but was separated by the replication region of the plasmid. Homologous recombination between the two direct repeats of IS1216V resulted in excision of the replication region of the plasmid to regenerate MESPM1. The p4780-1 and pV19 of E. faecium carried MES6272-2-like structures with IS1216V, albeit with multiple insertions by other insertion sequences. The findings show that IS1216V plays important roles in bidirectional gene transfer of multidrug resistance between enterococci and S. aureus.
Keywords: Staphylococcus aureus, enterococci, IS1216V, gene transfer
1. Introduction
Sequence type 59 (ST59) methicillin-resistant Staphylococcus aureus (MRSA) is the dominant type of community-associated MRSA (CA-MRSA) in Taiwan and can be divided into two clones: Panton–Valentine leukocidin (PVL)-positive/staphylococcal cassette chromosome mec (SCCmec) V and PVL-negative/SCCmec IV [1,2,3]. Previously, we reported that ST59 S. aureus acquires IS1216V-mediated enterococcal composite transposons MES, which are responsible for multidrug resistance [4,5]. ST59 PVL-positive/SCCmec V MRSA acquires MESPM1, which confers resistances to erythromycin, kanamycin, streptomycin, and chloramphenicol; ST59 PVL-negative/SCCmec IV MRSA usually acquires MES6272-2, which confers resistances to erythromycin, kanamycin, gentamicin, and chloramphenicol. Both the MESPM1 and MES6272-2 are inserted into the chromosomal sasK gene with an 8 bp att sequence and are flanked by direct repeats of IS1216V at both ends, indicating that MESPM1 and MES6272-2 are composite transposons mediated by IS1216V [4,5]. IS1216V, belonging to the IS6/IS26 family, is 809 bp in length with 18 inverted repeats [6,7]. Although IS1216V is an enterococcal insertion sequence rarely found in S. aureus, up to five copies of IS1216V are located in MESPM1 and MES6272-2 of ST59 S. aureus [6].
Regarding multidrug resistance in ST59 S. aureus, four resistance determinants originate in enterococci, including ermB (erythromycin resistance), aph(3′)-IIIa (kanamycin resistance), aadE (streptomycin resistance), and aacA-aphD (gentamicin resistance), while the cat gene (chloramphenicol resistance) and its surrounding genetic environment is native to S. aureus. MESPM1 includes ermB, aph(3′)-IIIa, aadE, and cat, while MES6272-2 includes ermB, aph(3′)-IIIa, truncated aadE, aacA-aphD, and cat. On the left side of MESPM1, the IS1216V-ermB-[aph(3′)-IIIa]-aadE cluster displays 100% DNA sequence similarity to the corresponding region in pLG2 of Enterococcus faecalis [5]. On the right side of MESPM1, the cat gene and its surrounding environment show high DNA sequence similarity (>99.5%) to the corresponding region of the SAP084A plasmid in S. aureus, albeit with four copies of IS1216V nearby [5]. The genetic organization of MES6272-2 is similar to that of MESPM1 except with disruption of the aadE gene by aacA-aphD and replacement of the region downstream of the aadE gene by group II introns [4].
E. faecalis and Enterococcus faecium are frequently encountered multidrug-resistant microorganisms and have rapidly emerged as troublesome pathogens causing nosocomial infections [8,9,10,11]. Mobile genetic elements (MGEs), including plasmids and transposons, have been shown to play a major role in dissemination of antibiotic resistance among the Enterococcus species [12,13]. Some enterococcal MGEs have even been shown to be transferrable to other pathogenic species, such as Staphylococcus aureus. Tn1546, which encodes vancomycin resistance via vanA, originates in enterococci and can transfer to S. aureus, leading to emergence of vancomycin-resistant S. aureus (VRSA) [12,14]. The structure of Tn1546 displays heterogeneity, such as an enterococcal insertion sequence IS1216V inserted into the backbone [15]. IS1216V is also considered to be part of a large mobile element containing Tn1546 and may cotransfer with Tn1546 to other species [16].
MESPM1 and MES6272-2 in ST59 MRSA, the composite transposons mediated by IS1216V, are examples of insertion sequences playing a pivotal role in the dissemination of antibiotic resistance. DNA sequence analysis revealed that more than half of the structures originate in enterococci, not in staphylococci. In the current study, we characterized enterococcal clinical isolates with MES-like structures and found evidence of IS1216V-mediated interspecies transfer of multidrug resistance genes between staphylococci and enterococci.
2. Materials and Methods
2.1. Bacterial Isolates
A total of 270 enterococcal isolates collected from clinical specimens were included in this study: (i) 95 randomly selected isolates collected between 1991 and 1993 at the National Taiwan University Hospital (NTUH), (ii) 26 randomly selected isolates from blood cultures collected between 2002 and 2003 at NTUH, and (iii) 149 isolates collected from January 2013 to October 2014 at the Kaohsiung Medical University Hospital (KMUH). Among the 149 isolates, E1–E51 (n = 50) were randomly selected vancomycin-susceptible enterococci collected from blood cultures, while V1–V99 (n = 99) were first identified as vancomycin-resistant enterococci and collected within the period. Enterococcal multiplex PCR or groEL sequencing was used for species identification [17]. Detailed information about the bacterial strains is shown in Table S1.
2.2. PCR Mapping of MES Structures
The resistance determinants ermB, aph(3′)-IIIa, aadE, aacA-aphD, and cat were detected as previously described [4]. For ermB-, aph(3′)-IIIa-, and aadE-positive isolates, six primer sets (I–VI) were used to screen the MES structures. The entire region of MESPM1-like structures in the two E. faecalis strains was further mapped using two PCR primer sets (A and B) with the methods we had previously described [5]. Information about the primers is indicated in Table 1, and the locations of the primers are indicated in Figure S1.
Table 1.
Primers used in this study.
| Primer Set | Primer Name | Sequence (5′ to 3′) | PCR Product Size (bp) |
|---|---|---|---|
| I | IS1216VF | AGTTTACGCACTGCCTCT | 2170 |
| tnpF | CGGTATCCTGGGTGT | ||
| II | IS1216V-fo | CTTCGGTTCATCAAACTGC | 1384 |
| tnp-rev | TCAAATCACCTTCCTACTACCC | ||
| III | tnp-fo | GCGTGTATCTTCGGAGGTA | 2727 |
| ermB-rev | TTGGAACAGGTAAAGGGC | ||
| IV | ermB-fo | ATCTGTGGTATGGCGGGTA | 1432 |
| aphIIIa-rev | ATGACATTGCCTTCTGCG | ||
| V | aphIIIa-fo | TGTCATACCACTTGTCCGC | 1345 (2331 if ISEfm1 insertion) |
| aadE-rev | GCTGCCTGGATAGCACATA | ||
| VI | aadE-R | GTTCCCGCCTCTCTTCTA | 2254 |
| aacA-aphD-F | ATACAGAGCCTTGGGAAG | ||
| A | 1F | AGTAGCCTTTCCCTCACTT | 1301 |
| 35R | GCTTTGACGCTATGACGA | ||
| B | 35F | CCTTACCAGTTGTTCCGAA | 1619 |
| LA-R2 | CCCATGCAGGTTTCAAAATGTGTAAGTCA |
2.3. Antimicrobial Susceptibility Testing
Susceptibility testing of vancomycin, erythromycin, kanamycin, streptomycin, gentamicin, and chloramphenicol was performed by the agar dilution method according to the 2020 guidelines of the Clinical and Laboratory Standards Institute [18]. E. faecalis ATCC 29,212 was used as the reference strain. High-level resistance of streptomycin and gentamicin was defined as minimal inhibitory concentrations of >2000 and >500 mg/L, respectively.
2.4. Multilocus Sequence Typing (MLST)
MLST of the 11 MES-carrying enterococcal isolates (2 E. faecalis and 11 E. faecium) was performed as previously described [19,20]. Sequence types (STs) were assigned according to the program on the MLST website (https://pubmlst.org). Clonal complex (CC) was defined by geoBURST [21].
2.5. Pulsed-Field Gel Electrophoresis (PFGE)
PFGE was performed as previously described [22]. In brief, agarose-embedded bacterial DNA was digested with SmaI (New England BioLabs, Ipswich, MA, USA) and then was separated using a CHEF-DRIII apparatus (Bio-Rad Laboratories, Hercules, CA, USA). PFGE was carried out at 200 V and 14 °C for 22.5 h, with the pulse times ranging from 5 to 35 s. The pulsotypes were analyzed by BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The dendrogram of pulsotype relationships was produced by the unweighted pair group method using arithmetic averages (UPGMA) based on Dice similarity indices.
2.6. S1 Nuclease-Digested PFGE
S1 nuclease-digested PFGE analysis was performed as previously described to determine the sizes of the plasmids [23]. The agarose-embedded bacterial DNA was incubated at 37 °C for 45 min with 10 units of Aspergillus oryzae S1 nuclease (Invitrogen). The reaction was stopped by transferring the agarose-embedded bacterial DNA to ES buffer (0.5 mM EDTA, 1% (w/v) Sarkosyl, pH 9.0) at 4 °C for 10 min. The plugs were applied to wells of 1.2% (w/v) agarose gels (Bio-Rad) and run in a CHEF-DRIII apparatus (Bio-Rad) with a pulse angle of 120° and pulse times of 45 s for 14 h and 25 s for 6 h at 200 V in 0.5× Tris-borate-EDTA (TBE). Each band was considered a unit length linear plasmid.
2.7. Southern Blot
The linear plasmids in the gel separated by S1-PFGE were depurinated, denatured, neutralized, and transferred to a Hybond-N+ nylon membrane (GE Healthcare, Chicago, IL, USA) by vacuum-blotting. Hybridization was achieved using the digoxigenin-labeled DNA probe specific for the aadE gene generated by PCR. The detection of hybridization was performed using an alkaline-phosphatase-conjugated anti-digoxigenin antibody (Roche Diagnostics GmbH, Mannheim, Germany) and the substrate CSPD (Roche Diagnostics GmbH) according to the manufacturer’s instructions.
2.8. Sequencing and Analysis of the MES Structures and Their Adjacent Environments
The sequence of the MES6272-2-like structure in E. faecium 4780-1 was determined using the primer sets for PCR mapping described above, and extension of the IS12126V upstream sequence was achieved by inverse PCR. For E. faecalis strain N48 and E. faecium V19, Illumina MiSeq 300 bp paired-end sequencing was used to determine the MESPM1-like and MES6272-2-like structures. Total Illumina sequencing output corresponded to approximately 146- and 301-folds of the N48 and V19 genome size, respectively. Contigs were obtained using de novo assembly with SPAdes v3.10.1 [24]. A total of 92 and 261 contigs were yielded for the N48 and V19 genomes, respectively. The gaps between the contigs were filled up by Sanger sequencing. The open reading frame (ORF) was determined using BLAST. The plasmid family was determined using PlasmidFinder 2.0 [25].
2.9. Filter Mating
Filter mating was carried out on BHI agar using E. faecalis N48 or E. faecium V19 as donors, and E. faecalis JH2-2 or S. aureus RN2677 as recipients. The donor–recipient mix cultures were placed on a filter with incubation at 37 °C for 24 h, followed by resuspension and shaking in BHI broth with incubation at 37 °C for 1 h. The cells were collected and plated on BHI agar containing 250 mg/L erythromycin, 100 mg/L rifampin, 100 mg/L streptomycin (if donor was E. faecalis N48), 100 mg/L gentamicin (if donor was E. faecium V19), or 25 mg/L fusidic acid (if recipient was E. faecalis JH2-2).
2.10. Nucleotide Sequence Accession Numbers
The nucleotide sequences (Supplementary Data) of pEflis48, p4780-1, and pV8919 characterized in this study have been deposited in GenBank under accession numbers MT877066–MT877068.
3. Results
3.1. Molecular Characteristics of MESPM1- or MES6272-2-Carrying Enterococcal Strains
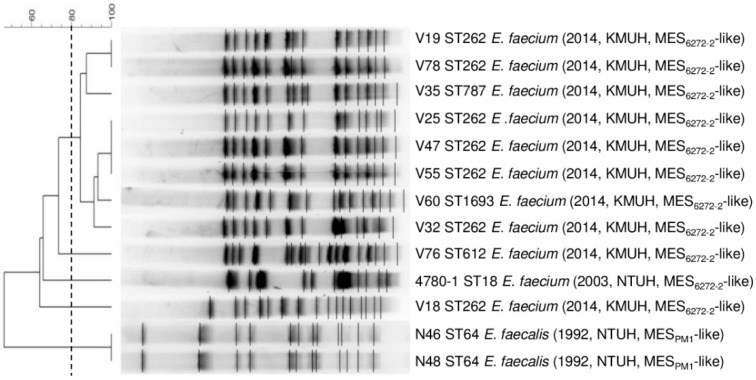
The 270 enterococcal isolates were first screened by PCR for five resistance determinants (ermB, aph(3′)-IIIa, aadE, aacA-aphD, and cat) and antimicrobial susceptibility testing (Table S1). A total of 128 isolates that were positive for ermB, aph(3′)-IIIa, and aadE by PCR were selected for further analysis because MESPM1 carried the ermB, aph(3′)-IIIa, and aadE, while MES6272-2 carried ermB, aph(3′)-IIIa, and the nearly complete aadE gene with 5′-end disruption by aacA-aphD. The PCR mapping of MES structures revealed that two E. faecalis strains isolated in 1992 harbored a MESPM1-like structure; one E. faecium strain isolated in 2003 and 10 E. faecium strains isolated from 2013 to 2014 harbored MES6272-2-like structures (Table 2). The phylogenetic relatedness of the 13 strains was analyzed by PFGE and MLST (Figure 1). The two E. faecalis strains belonged to ST64 and had identical SmaI PFGE patterns. For the 11 E. faecium strains, all of them belonged to clonal complex 17 (CC17) with 5 different STs (ST18, ST262, ST612, ST787, and ST1693). The E. faecium strains were all resistant to vancomycin except strain 4780-1 (Table S1). The eight strains were clustered together, distant from V18, V76, and 4780-1 (the only strain isolated in 2003).
Table 2.
Summary of the bacterial strains collected in this study.
| No. of Isolates | ||||
|---|---|---|---|---|
| Isolation Year | Total | |||
| 1991–1993 | 2002–2003 | 2013–2014 | ||
| Enterococcus species | 95 | 26 | 149 | 270 |
| E. faecalis | 82 | 14 | 30 | 126 |
| E. faecium | 10 | 12 | 117 | 139 |
| Others | 3 (E. hirae) | 0 | 2 (E. raffinosus) | 5 |
| Positive for ermB+, aph3′-IIIa+, aadE+ | 54 | 11 | 63 | 128 |
| MES structures | ||||
| MESPM1-like | 2 (E. faecalis) | 0 | 0 | 2 |
| MES6272-2-like | 0 | 1 (E. faecium) | 10 (E. faecium) | 11 |
Figure 1.
PFGE analysis of the 13 MES-carrying enterococcal strains. The dendrogram was produced by BioNumerics software, showing distance calculated by the Dice similarity index of SmaI-digested DNA fragments. The degree of similarity is shown in the scale. The strain number, ST, species, isolation year, isolation hospital, and MES types are indicated in the figure.
3.2. Localization of the MESPM1- or MES6272-2-Like Elements in Enterococcal Strains
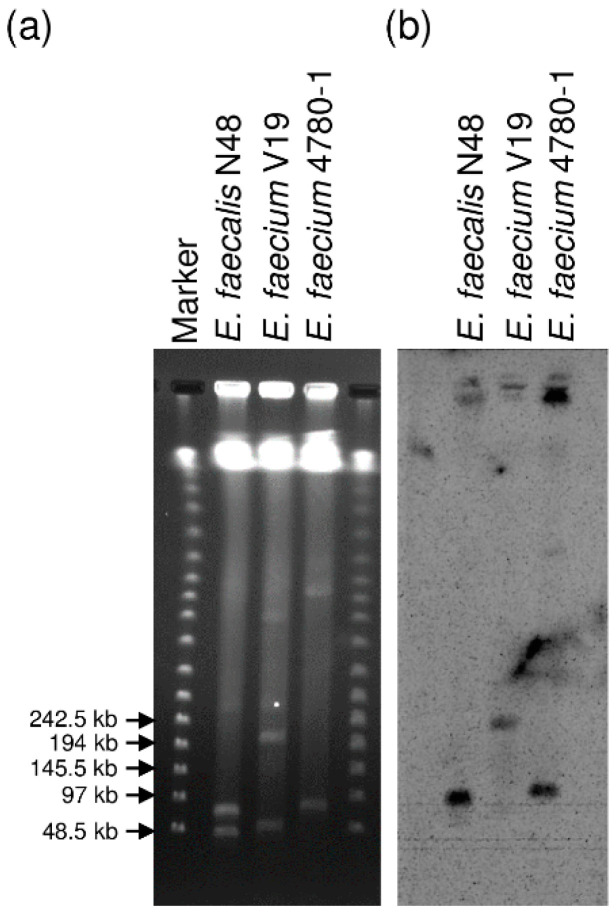
To determine the location of the MESPM1- or MES6272-2-like elements in enterococcal strains, E. faecalis strain N48 isolated in 1992, E. faecium 4780-1 isolated in 2003, and E. faecium V19 isolated in 2013 were chosen for S1 nuclease-digested PFGE analysis. The S1 nuclease-digested DNAs were separated by PFGE, followed by Southern blot hybridization with the aadE probe (Figure 2). Southern blot hybridization after S1 nuclease-digested PFGE revealed that the MES-like elements of E. faecalis strain N48 and E. faecium 4780-1 were located on the plasmids with estimated sizes ranging from 48.5 to 97 kb, while the MES6272-2-like elements of E. faecium V19 were located on a relatively large plasmid with an estimated size ranging from 194 to 242.5 kb. The size of the MESPM1-like-carrying plasmid of E. faecalis strain 46 and the MES6272-2-like-carrying plasmids of E. faecium strains V32, V35, V47, and V76 corresponded to E. faecalis N48 and E. faecium V19, respectively (data not shown).
Figure 2.
S1 nuclease PFGE analysis followed by Southern blot hybridization with the aadE probe of E. faecalis N48, E. faecium V19, and E. faecium 4780-1: (a) S1 nuclease-digested DNAs separated by PFGE; (b) Southern blot hybridization of the aadE probe of the S1 nuclease-digested DNAs.
3.3. Sequencing of the MESPM1-Like Elements and the Adjacent Genetic Environments in Enterococcal Strains
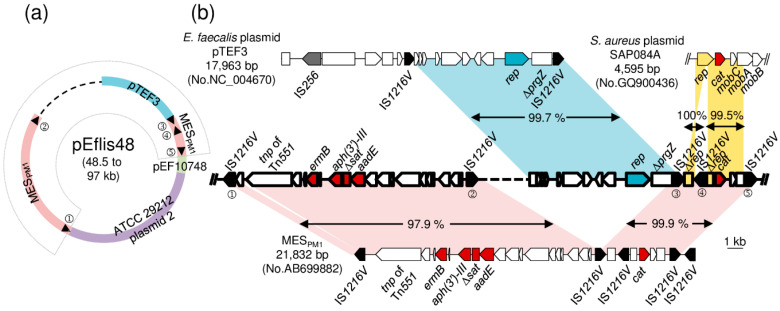
E. faecalis N48 was chosen for DNA sequencing to determine the MES-like structure and the surrounding genetic environment. An incomplete plasmid sequence of 50,564 bp in length was determined, and sequence analysis revealed that the MESPM1-like structure was located on a mosaic plasmid, pEflis48 (Figure 3a). Positions 1–9944 nt showed 99.7% DNA sequence identity to the corresponding region of the E. faecalis plasmid pTEF3. Positions 9136–14,615 and 34,166–50,564 showed 98.4% DNA sequence identity to MESPM1 in ST59 MRSA. Positions 14,616–16,723 and 16,724–34,144 showed 99.3% and 99.0% DNA sequence identity to the corresponding regions of the E. faecalis plasmid pEF10748 and E. faecalis ATCC 29,212 plasmid 2, respectively.
Figure 3.
Structure of pEflis48 in E. faecalis N48. (a) Schematic map of pEflis48 is shown. Blue, pink, green, and purple colors in the circle indicate homologous regions to E. faecalis plasmid pTEF3 (GenBank accession number NC_004670), ST59 MRSA MESPM1 (GenBank accession number AB699882), E. faecalis plasmid pEF10748 (GenBank accession number MK993385), and E. faecalis ATCC 29,212 plasmid 2 (GenBank accession number CP008814). The black triangle indicates the position of IS1216V. The dashed line in the circle indicates an unresolved sequence. (b) Genetic organization of the fan-shaped region indicated in (a) is shown. The arrow indicates an open reading frame. The rectangle indicates a truncated gene. Black, grey, red, and blue arrows indicate tnp (transposase) of IS1216V, tnp of IS256, resistance determinates, and rep (replication initiator) gene, respectively. Homologous regions to E. faecalis plasmid pTEF3, S. aureus plasmid SAP084A, and MESPM1 are shaded in blue, yellow, and red, respectively.
Detailed sequence comparison of the MESPM1-like structure in E. faecalis N48 is shown in Figure 3b, and the results are highly identical to those for ST59/SCCmec V MRSA PM1, except (i) an additional 344 bp sequence near the IS1216V①; (ii) an additional region corresponding to E. faecalis plasmid pTEF3, including a rep gene belonging to the Inc18 family of broad-host-range conjugative plasmid, a truncated gene-encoding pheromone-binding protein, and a copy of IS1216V; and (iii) lack of a 298 bp noncoding sequence and an IS1216V on the right side of MESPM1. Interestingly, the cat gene and its surrounding environment that originated in S. aureus were also found in pEflis48 and showed 99.9% sequence identity to MESPM1. Moreover, PCR and DNA sequencing using the primer set A located in MESPM1 (one was located upstream of IS1216V② and the other was located downstream of IS1216V③, as indicated in Figure 3 and Figure S1) outside the pTEF3 region yielded a 1301 bp amplicon corresponding to a copy of IS1216V, indicating that homologous recombination occurred between the two direct repeats of IS1216V to excise the pTEF3 region. PCR of E. faecalis N46 generated the identical size to that of E. faecalis N48 (Figure S1b).
3.4. Sequencing of the MES6272-2-Like Elements and the Adjacent Genetic Environments in Enterococcal Strains
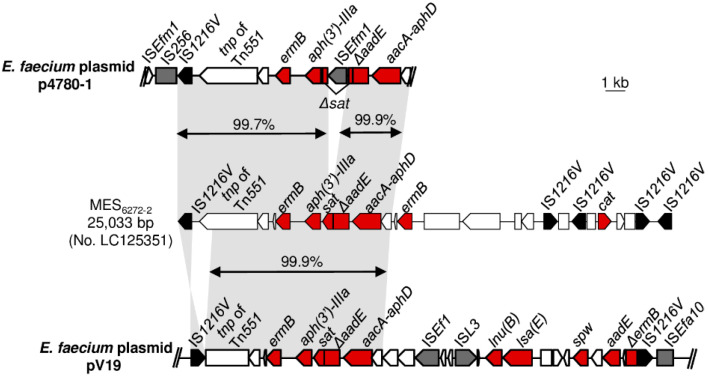
E. faecium 4780-1 and V19 both carried MES6272-2-like structures. Sequence analysis revealed that the IS1216V-ermB-[aph(3′)-IIIa]-sat-ΔaadE-[aacA-aphD] cluster of the MES6272-2-like structure in E. faecium 4780-1 showed >99.7% identity to the corresponding region in ST59/SCCmec IV MRSA 6272-2, except an ISEfm1 insertion into the sat gene (Figure 4).
Figure 4.
Genetic organization of MES6272-2-like structures in p4780-1 and pV19. The arrow indicates an open reading frame. The rectangle indicates a truncated gene. Black, grey, and red colors indicate tnp (transposase) of IS1216V, tnp of other insertion sequences, and resistance determinates, respectively. Homologous regions are shaded.
For the MES6272-2-like structures of E. faecium V19, Illumina sequencing revealed that the MES6272-2-like structure was located on plasmid, and 24,402 bp in length was determined. The ermB-[aph(3′)-IIIa]-sat-ΔaadE-[aacA-aphD] cluster of MES6272-2-like structures in E. faecium V19 showed 99.9% identity to the corresponding region in ST59/SCCmec IV MRSA 6272-2. However, the 3′-end of transposase of Tn551 was absent in E. faecium V19, and the direction of IS1216V in front of ermB was different between E. faecium V19 and ST59/SCCmec IV MRSA 6272-2. The region between the aacA-aphD gene and the second IS1216V of E. faecium V19 displayed different structures compared with MES6272-2, harboring multiple resistance determinants of lnu(B) (lincosamide resistance), lsa(E) (pleuromutilin, lincosamide, and streptogramin A resistance), spw (spectinomycin resistance), and an aadE gene.
3.5. Horizontal Transfer of MESPM1 or MES6272-2 Encoding Drug Resistance
Conjugative transfer of MES-like structures between enterococci and S. aureus was performed by filter mating. E. faecalis N48 or E. faecium V19 were used as donors, while E. faecalis JH2-2 or S. aureus RN2677 were used as recipients. The MES-related resistances were successfully transferred from E. faecalis N48 or E. faecium V19 to E. faecalis JH2-2, with transfer frequencies of 3.7 × 10−2 and 2.5 × 10−7, respectively. Resistances were transferred to transconjugants (Table 3). All of the transconjugants tested acquired plasmids with sizes similar to their donors detected by S1 nuclease PFGE with Southern blot hybridization. However, no transconjugants were obtained in the cases of transfer from E. faecalis N48 or E. faecium V19 to S. aureus RN2677 (efficiency < 6.5 × 10−10).
Table 3.
Transfer of drug resistance in filter mating.
| Transfer Frequency | MIC (mg/L) | |||||
|---|---|---|---|---|---|---|
| E | KM | SM | GM | C | ||
| Recipient: E. faecalis JH2-2 | 0.25 | 64 | 128 | 16 | 8 | |
| Donor: E. faecalis N48 | 3.7 × 10−2 | >256 | >1024 | >1024 | >1024 | 64 |
| Transconjugants (E. faecalis N48) | >256 | >1024 | >1024 | - | 64 | |
| Donor: E. faecium V19 | 2.5 × 10−7 | >256 | >1024 | 32 | >1024 | 8 |
| Transconjugants (E. faecium V19) | >256 | >1024 | - | 512 | - | |
Abbreviation: E, erythromycin; KM, kanamycin; SM, streptomycin; GM, gentamicin; C, chloramphenicol. “-”, not detected.
4. Discussion
Previously, we described MESPM1 and MES6272-2 in ST59 MRSA, the novel multidrug-resistant composite transposons mediated by enterococcal IS1216V and mingled with S. aureus- and enterococci-originated sequences [4,5]. This raised a question about how the mosaic composite transposons emerged and disseminated into ST59 MRSA, the dominant CA-MRSA in Taiwan identified in 1997 [26]. In the current study, we collected enterococcal clinical isolates from as early as 1991 and analyzed their resistance elements. As expected, MESPM1-like and MES6272-2-like structures, both surrounded by IS1216V, were found in E. faecalis and E. faecium isolates, respectively. Furthermore, the MESPM1-like structure of E. faecalis isolated in 1992 had already acquired the cat gene and its surrounding genetic environment that was native to S. aureus, suggesting that horizontal gene transfer from S. aureus to enterococci might have occurred before the emergence of multidrug-resistant ST59 MRSA.
The IS1216 isoform groups, including IS1216, IS1216E, and IS1216V, are one of the most multiply represented insertion sequences in enterococci and play important roles in the dissemination of resistance determinants [7,12]. They form composite transposons encoding antibiotic resistance in enterococci, such as Tn5385 (encoding gentamicin, streptomycin, penicillin, erythromycin, and tetracycline/minocycline resistances), Tn5482, and Tn5506 (both encoding vancomycin resistance) [27,28,29]. MESPM1 and MES6272-2 are rare cases of composite transposons mediated by IS1216V found in S. aureus [4,5]. We previously reported that MESPM1 would be excised, resulting in a single IS1216V remaining in MESPM1 [4]. The same phenomenon has been demonstrated in pEf37BA of E. faecium with the pbp5 gene surrounded by two direct repeats of IS1216V [30]. In the current study, the pEflis48 of ST64 E. faecalis carried a MESPM1-like structure, within which a region included the rep gene and a truncated prgZ gene encoding pheromone-binding proteins corresponding to that in pTEF3 of vancomycin-resistant E. faecalis (Figure 3). PCR with primer set A located upstream of IS1216V② and downstream of IS1216V③ generated a 1031 bp amplicon, indicating that RecA-mediated homologous recombination would occur to delete the replication region of pTEF3 and leave a copy of IS1216V (Figure 3 and Figure S1). As a result, MESPM1, a composite transposon mediated by IS1216V highly similar to that in ST59/SCCmec V MRSA with erythromycin, kanamycin, streptomycin, and chloramphenicol resistances, would be regenerated. This indicates the important role of IS1216V in the dissemination of multidrug resistance between enterococci and ST59 S. aureus, the most dominant CA-MRSA in Taiwan.
The cat genes found in staphylococci and enterococci are frequently located on small multicopy plasmids and are seldom associated with large multiresistant plasmids [31,32]. In pEflis48 of E. faecalis and MESPM1 of ST59/SCCmecV MRSA, the cat gene and its surrounding environments were both highly related to a small S. aureus plasmid SAP084A (>99.5% DNA sequence similarity, Figure 3), which indicates that this region originated in S. aureus. Furthermore, the S. aureus-originated cat region in pEflis48 and MESPM1 was flanked by two direct repeats of enterococcal IS1216V in which another copy embedded, resulting in disruption of the rep gene encoding replication initiator of the S. aureus plasmid. This implies that the region transferred from S. aureus to enterococci, mediated by enterococcal IS1216V. Therefore, we hypothesize that the S. aureus-originated cat region was initially acquired on a large multiresistant plasmid of enterococci by IS1216V from a small S. aureus plasmid, regenerated to MESPM1 by RecA-mediated homologous recombination via two direct repeats of IS1216V, and finally disseminated into ST59 S. aureus, the dominant CA-MRSA in Taiwan. This would be the first example of IS1216V-mediated bidirectional transfer of resistance in S. aureus and enterococci.
The MESPM1-like structure of E. faecalis N48 was located on an Inc18 plasmid. The Inc18 plasmids usually possess the ability to confer antibiotic resistance and to transfer to a variety of Gram-positive cocci including enterococci and staphylococci [12,33]. Inc18 plasmids comprising insertion of Tn1546 to facilitate dissemination of vancomycin resistance from enterococci to MRSA have been reported [34]. In the current study, although pEflis48 displayed high transfer efficiency (3.7 × 10−2/donor) from donor E. faecalis N48 to recipient E. faecalis JH2-2, it failed to transfer to S. aureus RN2677. The reason is unknown and needs to be investigated in the future. A previous study reported that a pSK41-like plasmid is necessary to contribute to successful transfer of the Inc18-like vanA plasmid from E. faecalis to MRSA [35]. Further research should be carried out to illustrate the transfer mechanism of MES-like structures between enterococci and S. aureus.
ST64 E. faecalis belongs to CC8. Although CC8 was not included in the major global clusters associated with healthcare-associated infections such as CC2 [8,9], ST64 E. faecalis has been reported to be associated with multidrug resistance to erythromycin, kanamycin, streptomycin, gentamicin, and tetracycline [36,37]. In contrast to E. faecalis, all of the E. faecium isolates characterized in this study were CC17, the most prevalent cluster worldwide, notably in the nosocomial setting [8,38,39]. It has been proposed that progressive evolution to acquire resistance determinants in CC17 E. faecium is associated with its success to adapt to conditions of modern hospitals [38]. In the current study, the MES6272-2-like structures of the CC17 E. faecium strains isolated in 2003, 2013, and 2014 were inserted by several insertion sequences such as ISEfm1, ISEf1, and ISL3, and an additional cluster of resistance determinants lnu(B), lsa(E), spw, and aadE was acquired (Figure 4). The unpredictable genomic changes of CC17 E. faecium would hamper the tracking of the origin of MES-like structures.
In conclusion, the MESPM1 structure with the enterococci-originated region (erythromycin, kanamycin, and streptomycin resistances) and the S. aureus-originated region (chloramphenicol resistance) found in ST64 E. faecalis and ST59 S. aureus indicates that bidirectional gene transfer mediated by IS1216V can occur between enterococci and S. aureus. Our study is of great importance, as it is the first to demonstrate the role of IS1216V in interspecies transfer of multidrug resistance genes between enterococci and ST59 S. aureus.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/12/1905/s1, Figure S1: MESPM1, pEflis48, MES6272-2, p4780-1, and pV19, Table S1: Information of bacterial strains. Supplementary Data: The nucleotide sequences of pEflis48, p4780-1, and pV8919 characterized in this study.
Author Contributions
Conceptualization, Y.-T.L. and W.-C.H.; methodology, Y.-T.L., S.-P.T. and W.-C.H.; software, C.-C.C. and W.-C.H.; validation, Y.-T.L., C.-C.C., Y.-H.C. (You-Han Chen) and W.-C.H.; formal analysis, C.-C.C., Y.-H.C. (You-Han Chen) and W.-C.H.; investigation, C.-C.C., Y.-T.J. and W.-C.H.; resources, S.-P.T., Y.-T.J., Y.-H.C. (Yen-Hsu Chen), L.-J.T. and W.-C.H.; data curation, S.-P.T. and W.-C.H.; writing—original draft preparation, Y.-T.L., W.-W.H. and W.-C.H.; writing—review and editing, Y.-T.L., W.-W.H. and W.-C.H.; visualization, W.-C.H.; supervision, S.-P.T., W.-W.H. and W.-C.H.; project administration, W.-C.H.; funding acquisition, Y.-H.C. (Yen-Hsu Chen) and W.-C.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of Taiwan (MOST104-2320-B-037-029 and MOST107-2320-B-037-021-MY3), the Kaohsiung Medical University (KMU-M106003), and partially by a Kaohsiung Medical University Research Center Grant (KMU-TC109B02).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen C.J., Huang Y.C. New epidemiology of Staphylococcus aureus infection in Asia. Clin. Microbiol. Infect. 2014;20:605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y.C., Chen C.J. Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s. Int. J. Antimicrob. Agents. 2011;38:2–8. doi: 10.1016/j.ijantimicag.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Su Y.C., Hung W.W., Lin J.M., Chang C.C., Chen Y.H., Lai Y.L., Tseng S.P., Lu P.L., Yamamoto T., Teng L.J., et al. Tracking the evolution of the two successful CC59 methicillin-resistant Staphylococcus aureus clones in Taiwan: The divergence time of the two clades is estimated to be the 1980s. Int. J. Antimicrob. Agents. 2020:106047. doi: 10.1016/j.ijantimicag.2020.106047. [DOI] [PubMed] [Google Scholar]
- 4.Hung W.C., Wan T.W., Kuo Y.C., Yamamoto T., Tsai J.C., Lin Y.T., Hsueh P.R., Teng L.J. Molecular Evolutionary Pathways toward Two Successful Community-Associated but Multidrug-Resistant ST59 Methicillin-Resistant Staphylococcus aureus Lineages in Taiwan: Dynamic Modes of Mobile Genetic Element Salvages. PLoS ONE. 2016;11:e0162526. doi: 10.1371/journal.pone.0162526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung W.C., Takano T., Higuchi W., Iwao Y., Khokhlova O., Teng L.J., Yamamoto T. Comparative genomics of community-acquired ST59 methicillin-resistant Staphylococcus aureus in Taiwan: Novel mobile resistance structures with IS1216V. PLoS ONE. 2012;7:e46987. doi: 10.1371/journal.pone.0046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/MMBR.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmer C.J., Hall R.M. An analysis of the IS6/IS26 family of insertion sequences: Is it a single family? Microb. Genom. 2019;5 doi: 10.1099/mgen.0.000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias C.A., Murray B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Solache M., Rice L.B. The Enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019;32:e00058-00018. doi: 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos S., Silva V., Dapkevicius M.L.E., Igrejas G., Poeta P. Enterococci, from harmless bacteria to a pathogen. Microorganisms. 2020;8:1118. doi: 10.3390/microorganisms8081118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollenbeck B.L., Rice L.B. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence. 2012;3:421–433. doi: 10.4161/viru.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegstad K., Mikalsen T., Coque T.M., Werner G., Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 14.Haaber J., Penades J.R., Ingmer H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017;25:893–905. doi: 10.1016/j.tim.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Clark N.C., Weigel L.M., Patel J.B., Tenover F.C. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 2005;49:470–472. doi: 10.1128/AAC.49.1.470-472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sletvold H., Johnsen P.J., Wikmark O.G., Simonsen G.S., Sundsfjord A., Nielsen K.M. Tn1546 is part of a larger plasmid-encoded genetic unit horizontally disseminated among clonal Enterococcus faecium lineages. J. Antimicrob. Chemother. 2010;65:1894–1906. doi: 10.1093/jac/dkq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung W.W., Chen Y.H., Tseng S.P., Jao Y.T., Teng L.J., Hung W.C. Using groEL as the target for identification of Enterococcus faecium clades and 7 clinically relevant Enterococcus species. J. Microbiol. Immunol. Infect. 2019;52:255–264. doi: 10.1016/j.jmii.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 18.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI; Wayne, PA, USA: 2020. Clinical and Laboratory Standards Institute. [Google Scholar]
- 19.Homan W.L., Tribe D., Poznanski S., Li M., Hogg G., Spalburg E., Van Embden J.D., Willems R.J. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002;40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz-Garbajosa P., Bonten M.J., Robinson D.A., Top J., Nallapareddy S.R., Torres C., Coque T.M., Canton R., Baquero F., Murray B.E., et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006;44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francisco A.P., Bugalho M., Ramirez M., Carrico J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung W.C., Chen H.J., Lin Y.T., Tsai J.C., Chen C.W., Lu H.H., Tseng S.P., Jheng Y.Y., Leong K.H., Teng L.J. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PLoS ONE. 2015;10:e0143106. doi: 10.1371/journal.pone.0143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton B.M., Harding G.P., Zuccarelli A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 24.Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., Lapidus A., Prjibelsky A., Pyshkin A., Sirotkin A., Sirotkin Y. Assembling genomes and mini-metagenomes from highly chimeric reads; Proceedings of the Annual International Conference on Research in Computational Molecular Biology; Beijing, China. 7–10 April 2013; pp. 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., Moller Aarestrup F., Hasman H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle-Vavra S., Ereshefsky B., Wang C.C., Daum R.S. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 2005;43:4719–4730. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice L.B., Carias L.L. Transfer of Tn5385, a composite, multiresistance chromosomal element from Enterococcus faecalis. J. Bacteriol. 1998;180:714–721. doi: 10.1128/JB.180.3.714-721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handwerger S., Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1995;39:2446–2453. doi: 10.1128/AAC.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heaton M.P., Discotto L.F., Pucci M.J., Handwerger S. Mobilization of vancomycin resistance by transposon-mediated fusion of a VanA plasmid with an Enterococcus faecium sex pheromone-response plasmid. Gene. 1996;171:9–17. doi: 10.1016/0378-1119(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 30.Morroni G., Brenciani A., Litta-Mulondo A., Vignaroli C., Mangiaterra G., Fioriti S., Citterio B., Cirioni O., Giovanetti E., Biavasco F. Characterization of a new transferable MDR plasmid carrying the pbp5 gene from a clade B commensal Enterococcus faecium. J. Antimicrob. Chemother. 2019;74:843–850. doi: 10.1093/jac/dky549. [DOI] [PubMed] [Google Scholar]
- 31.Roberts M.C., Schwarz S. Antimicrobial Drug Resistance. Springer; Berlin/Heidelberg, Germany: 2017. Tetracycline and chloramphenicol resistance mechanisms; pp. 231–243. [Google Scholar]
- 32.Schwarz S., Kehrenberg C., Doublet B., Cloeckaert A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004;28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Kohler V., Vaishampayan A., Grohmann E. Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms. Plasmid. 2018;99:11–21. doi: 10.1016/j.plasmid.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhu W., Murray P.R., Huskins W.C., Jernigan J.A., McDonald L.C., Clark N.C., Anderson K.F., McDougal L.K., Hageman J.C., Olsen-Rasmussen M., et al. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:4314–4320. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W., Clark N., Patel J.B. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob. Agents Chemother. 2013;57:212–219. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinones D., Kobayashi N., Nagashima S. Molecular epidemiologic analysis of Enterococcus faecalis isolates in Cuba by multilocus sequence typing. Microb. Drug Resist. 2009;15:287–293. doi: 10.1089/mdr.2009.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo M., Nomura T., Yomoda S., Tanimoto K., Tomita H. Nosocomial infection caused by vancomycin-susceptible multidrug-resistant Enterococcus faecalis over a long period in a university hospital in Japan. Microbiol. Immunol. 2014;58:607–614. doi: 10.1111/1348-0421.12190. [DOI] [PubMed] [Google Scholar]
- 38.Lee T., Pang S., Abraham S., Coombs G.W. Antimicrobial-resistant CC17 Enterococcus faecium: The past, the present and the future. J. Glob. Antimicrob. Resist. 2019;16:36–47. doi: 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Leclercq R. Epidemiological and resistance issues in multidrug-resistant staphylococci and enterococci. Clin. Microbiol. Infect. 2009;15:224–231. doi: 10.1111/j.1469-0691.2009.02739.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.