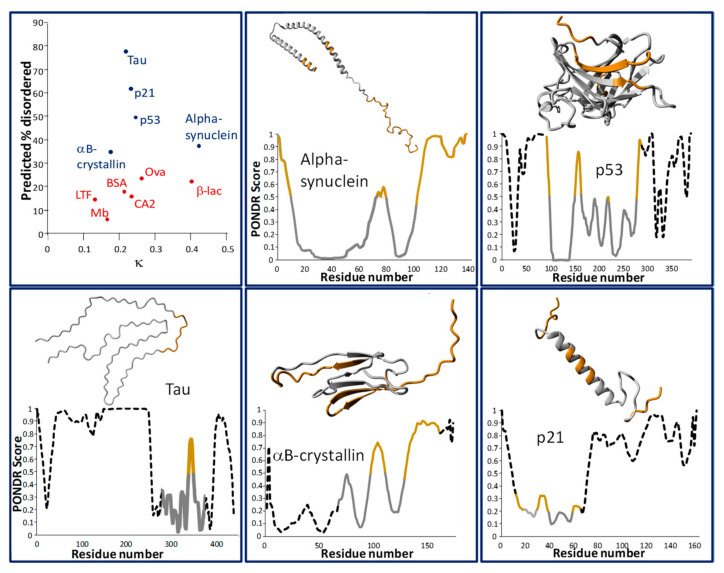
Figure 1.
(Top-left) Predicted fraction of residues in disordered regions versus charge mixing parameter κ for five intrinsically disordered (Tau, p21, p53, αB-crystallin, and alpha-synuclein) and six globular (myoglobin, lactotransferritin, carbonic anhydrase 2, bovine serum albumin, chicken ovalbumin, and bovine β-lactoglobulin) proteins. (Other panels) show results for the five intrinsically disordered proteins (IDPs) in detail, displaying (partial, except for alpha-synuclein) experimentally obtained structures and the PONDR (Predictor of Natural Disordered Regions) score per residue. Note that for several of these structures, a complex with interacting proteins or ligands (not shown) was analyzed rather than the monomeric IDP, resulting in a more ordered structure than might be expected. Regions predicted to be disordered by the PONDR algorithm (score > 0.5) are shown in gold in both the graphs and structures, except for p21, where a lower cut-off (0.2) was used to highlight the local maxima due to none of the parts of the sequence which are included in the experimentally observed structure scoring as “disordered”. PONDR scores for regions which are not part of the experimentally obtained structure are shown as a black dotted line in the graphs, regardless of their value. Protein Data Bank (PDB) identifiers for the structures are as follows—tau: 6TJO (441-residue isoform; structure obtained by cryo-EM of filaments); p21: 6P8H (XRD; measured as part of a complex with cyclin-dependent kinase 4 and cyclin D1); p53: 1TUP (XRD; measured as part of a complex with DNA); aB-crystallin: 3L1G (XRD); αSN: 1XQ8 (solution NMR; micelle-bound). Unless otherwise noted, the unmodified human sequence variant of these proteins was used in the calculations. Protein structure visualizations were generated using YASARA View [48].