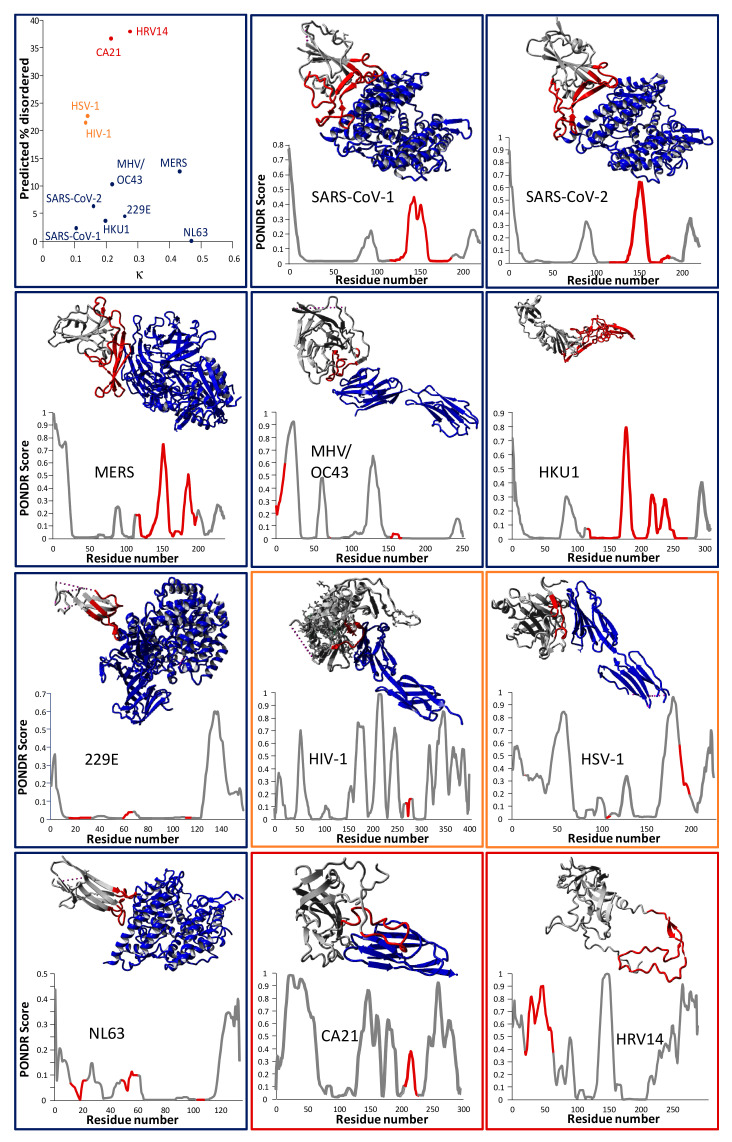
Figure 3.
(Top-left) The same analysis as in Figure 1 was performed for the key cell entry proteins of 11 human viruses, i.e., all seven human coronaviruses, HIV-1, HSV-1, CA21, and HRV14. Interestingly, the coronavirus spike proteins (in blue; targeting various receptors) are less disordered overall than the other four viral proteins, but span a range of κ values, while HIV-1/HSV-1 (orange; targeting CD4 [68] and nectin-1 [69], respectively) and CA21/HRV14 (red; both targeting ICAM-1) [70,71] seem to cluster together in the 2D plot. In the (other panels), crystal structures of the viral proteins are shown in the complex with their receptors (except for HCoV-HKU1 and HRV14, for which no structure of this complex was readily available), together with PONDR scores per residue. Border colors of these frames match the dots in the top-left panel. The receptor-binding domain (RBD) is shown in red in both the graphs and the structures. No crystal structure was readily available for the HCoV-OC43 spike protein. Therefore, the spike protein of the closely related (71% genome identity) murine hepatitis virus A59 (MHV) is shown [72]. PDB identifiers for the structures are as follows—SARS-CoV-1: 2AJF (XRD); SARS-CoV-2: 6LZG (XRD); CA21: 1Z7Z (cryo-EM); HCoV-NL63: 3KBH (XRD); MHV: 3R4D (XRD); HSV-1: 3SKU (XRD); MERS-CoV: 4L72 (XRD); HCoV-HKU1: 5KWB (XRD); HCoV-229E: 6ATK (XRD); HRV14: 1HRV (XRD); HIV-1: 1GC1 (XRD). Grey dashed lines present in some of the structures represent short stretches of the sequence which were not resolved. Protein structure visualizations were generated using YASARA View [48].