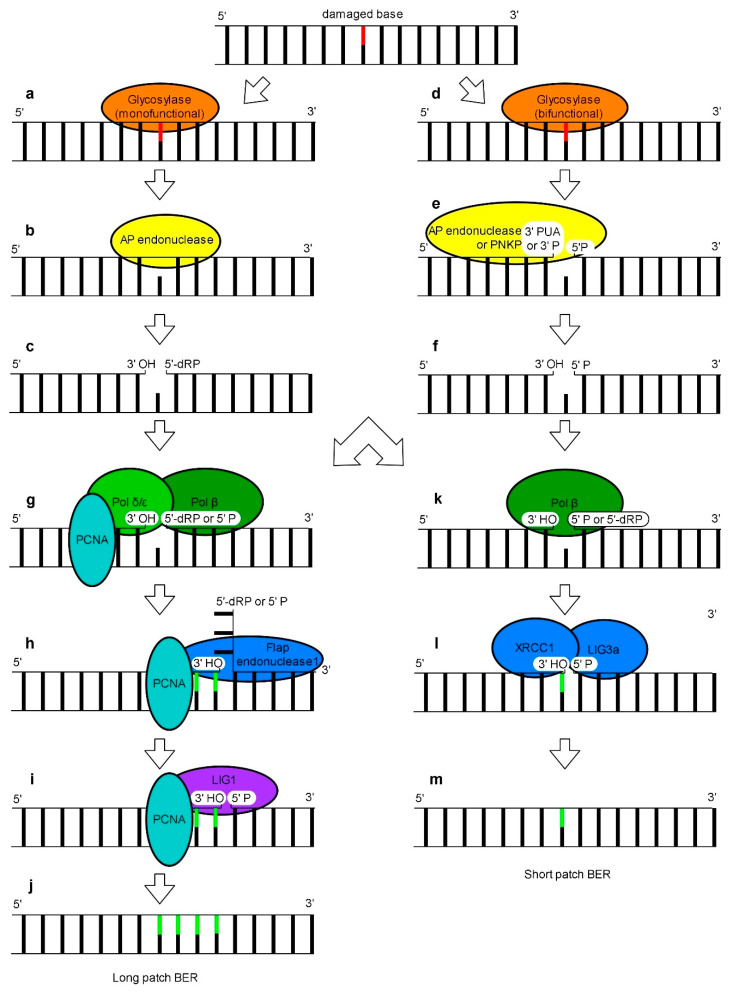
Figure 3.
A schematic representation of the long patch BER and short patch BER mechanism based on a mammalian model. In BER, the recognition and removal of a damaged base by a monofunctional glycosylase (a) results in the formation of an apurynic/apyrimidinic (AP) site (b). Next, an AP endonuclease cleaves a sugar backbone resulting in free 3′OH and 5′-dRP termini (c). In an alternative scenario, a bifunctional glycosylase (d) forms a Schiff base and cleaves a sugar backbone leading to the formation of 3’-PUA or 3’-P and 5’-P termini (e) followed by the conversion of 3’-PUA or 3’P to 3′OH by an AP endonuclease or phosphatase (PNKP (polynucleotide kinase 3’-phosphatase) in mammals), respectively (f). Products of reactions initiated by mono- and bifunctional glycosylases can undergo repair by the LP-BER (g–j) or SP-BER (k–m) subpathway. In LP-BER, where the conversion of 5′-dRP to 5′-P terminus is not necessary, pol β inserts the first nucleotide while the following ones are incorporated by pol δ and pol ε in cooperation with PCNA (g). Displaced nucleotides form a flap structure which is cleaved by FEN1 (h). Finally LIG1 ligates the adjacent 3’ and 5’ ends of a resynthesized and nicked DNA fragment (i). The repaired DNA has between 3-10 resynthesized nucleotides (j). In SP-BER, pol β converts 5’-dRP generated by a monofunctional glycosylase to 5’-P terminus. Moreover, it fills one nucleotide gap generated by an AP endonuclease or a bifunctional glycosylase in cooperation with an AP endonuclease (k). It is proposed that in plants pol λ may play the role of pol β. The XRCC1 and LIG3a complex ligates 3’ OH of an inserted nucleotide with 5’-P of a DNA backbone (l). As the result only one, damaged, nucleotide in a DNA strand is replaced (m).