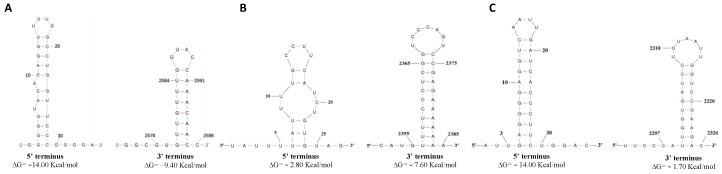
Figure 3.
Predicted secondary structures of the MOBV5, MOBV6, and MOBV7 terminal sequences. (A) The predicted secondary structures of the 5′- and 3′- terminal sequences of MOBV5. (B) The predicted secondary structures of the 5′- and 3′- terminal sequences of MOBV6. (C) The predicted secondary structures of the 5′- and 3′- terminal sequences of MOBV7. The RNAfold server was used for predicting the secondary structures of the termini, and for calculating the free energies as well. The 5′ and 3′-terminal sequences of each virus are folded to form potential stable stem-loop structures.