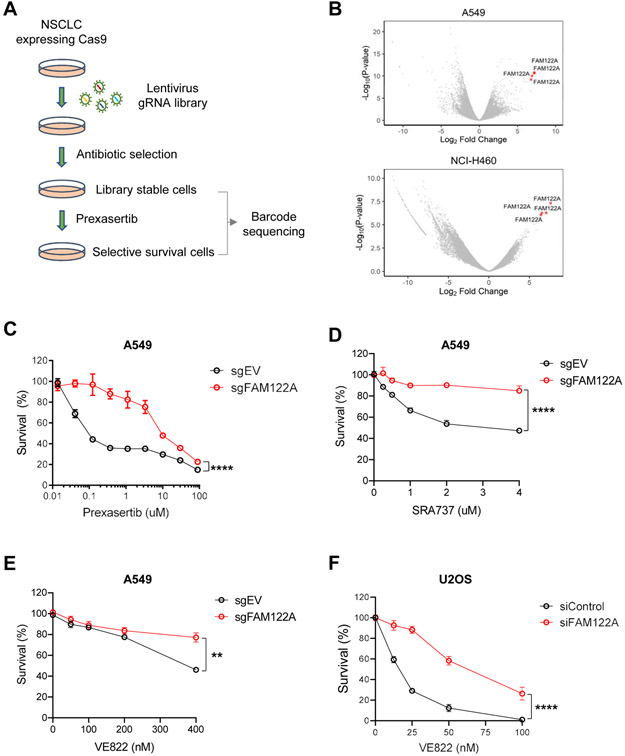
Figure 1. Genome-wide CRISPR screen reveals that FAM122A loss results in resistance to CHK1i and ATRi in lung cancer cells.
(A) Schematic of CRISPR-based screen for resistance to CHK1i (prexasertib) in lung cancer cells. Cells were infected with the Brunello sgRNA library (containing ~74,000 different sgRNAs) with a multiplicity of infection of 0.3. Following puromycin selection, cells were passaged for three weeks in the presence of prexasertib.
(B) Volcano plots showing genes targeted by sgRNAs that differentially dropped out or enriched following prexasertib selection in A549 and NCI-H460 cells. Note that all four gRNAs directed to FAM122A were found to be significantly enriched following prexasertib selection in A549 and NCI-H460 cells.
(C) Survival plots of control and FAM122A-KO A549 cells after exposure to CHK1i (prexasertib) for 5 days. Data are shown as mean ± SD from three independent experiments. ****P<0.0001, statistical analysis was performed using two-way ANOVA.
(D) Survival plots of control and FAM122A-KO A549 cells after exposure to CHK1i (SRA737) for 5 days. Data are shown as mean ± SD from three independent experiments. ****P<0.0001, statistical analysis was performed using two-way ANOVA.
(E) Survival plots of control and FAM122A-KO A549 cells after exposure to ATRi (VE822) for 5days. Data are shown as mean ± SD from three independent experiments. **P<0.01, statistical analysis was performed using two-way ANOVA.
(F) Survival plots of siControl and siFAM122A U2OS cells after exposure to ATRi (VE822) for 10days. Data are shown as mean ± SD from three independent experiments. ****P<0.0001, statistical analysis was performed using two-way ANOVA.