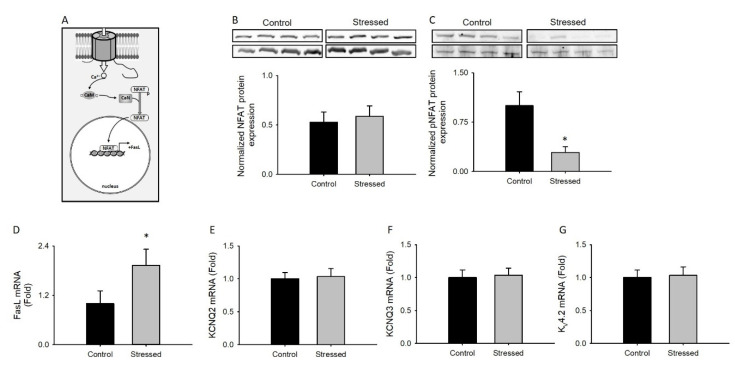
Figure 7.
NFAT-dependent signaling in CRS-subjected animals: (A) Schematic diagram depicting the signaling pathway studied in this figure. Bar graph of NFAT protein expression normalized to α-tubulin (B, n = 8) or pNFAT normalized to the total NFAT protein (C, n = 8) from whole hippocampus lysates; in the upper part of each graph, a representative immunoblot is shown. Bar graph of semi-quantitative PCR expression of genes known to be controlled by NFAT such as FasL mRNA (D), KCNQ2 mRNA (E), KCNQ3 mRNA (F), or Kv4.2 mRNA (G) isolated from whole hippocampus of juvenile rat controls (n = 8) or subjected to CRS for 3 weeks (n = 8). Bar graphs are mean ± SEM; black bars are control animals, and gray bars correspond to CRS-subjected animals * p value < 0.05.