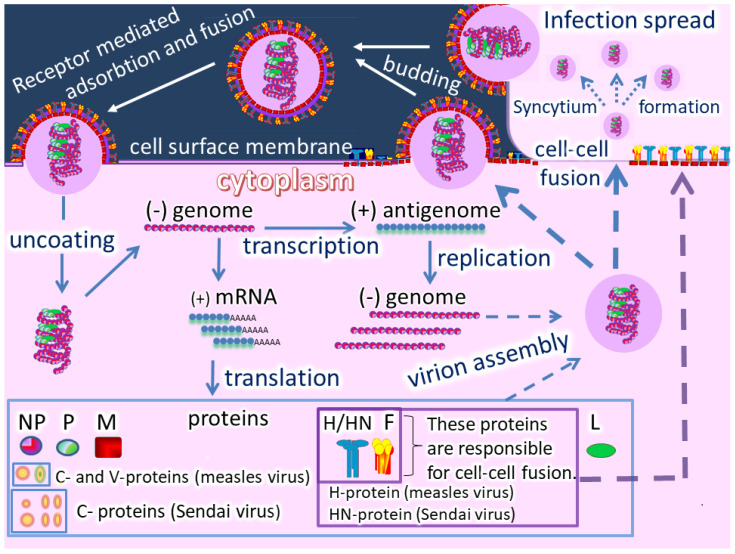
Figure 2.
A visual representation of the cell cycle of measles virus (MV) and Sendai virus (SeV). Both viruses belong to the Paramyxoviridae family, but MV belongs to the Morbillivirus genus and SeV to the Respirovirus genus. The life cycles of MV and SeV are very similar, but there are several important differences. Their attachment to host cells occurs through different cell entry receptors and different viral cell attachment proteins. The MV virus uses an H protein with hemagglutinin activity, while SeV uses the HN protein with hemagglutinin (H) and neuraminidase (N) activities. In addition to these proteins, the genomes of these viruses encode 5 structural proteins and accessary proteins. The main structural proteins for both viruses are: Nucleoprotein (N), Phosphoprotein (P), Matrix protein (M), Fusion protein (F), and Large Protein (L). The MV genome encodes two non-structural proteins, C and V, [20], while the SeV genome encodes a set of non-structural proteins, collectively referred as C-proteins (C’, C, Y1, Y2, V, W) [21]. Viral replication for MV and SeV follows a negative-stranded RNA virus replication model in which genomic RNA (minus strand) is used as a template to create a copy of positive sense RNA, employing the RNA-dependent RNA polymerase embedded in the virion. The plus RNA is further used as a template for making multiple copies of the minus RNA. The plus RNA is also translated by the host’s ribosomes, producing all viral proteins. Viruses are then assembled from these proteins along with genomic RNA and budded from the host cell. Both MV and SeV can form syncytia by fusing neighboring infected and non-infected cells into a polykayion.