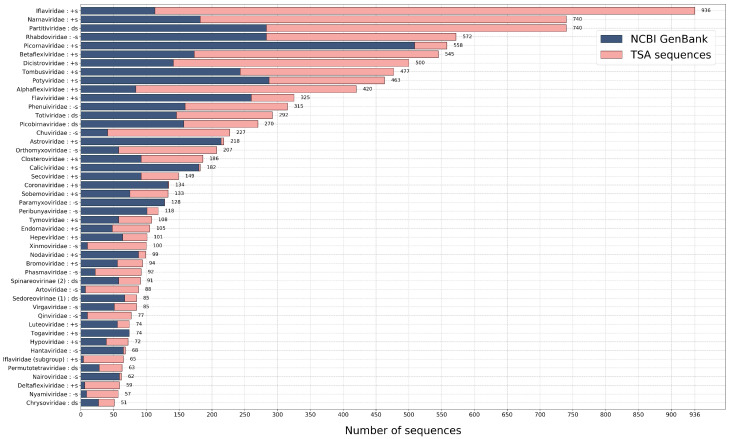
Figure 1.
The number of identified non-identical RdRp sequences. Each identified sequence with high score was trimmed to the RdRp core and then sorted into the genetically most similar group (the best HMM profile match). The 100% identical sequences within a group have been removed. Here, we only show groups, where the total number of sequences left are above 50. The genome type of each group is identified as follows: +s is positive single-stranded, -s is negative single-stranded and ds is double-stranded RNA genome. The source of the sequences is identified by different colours.