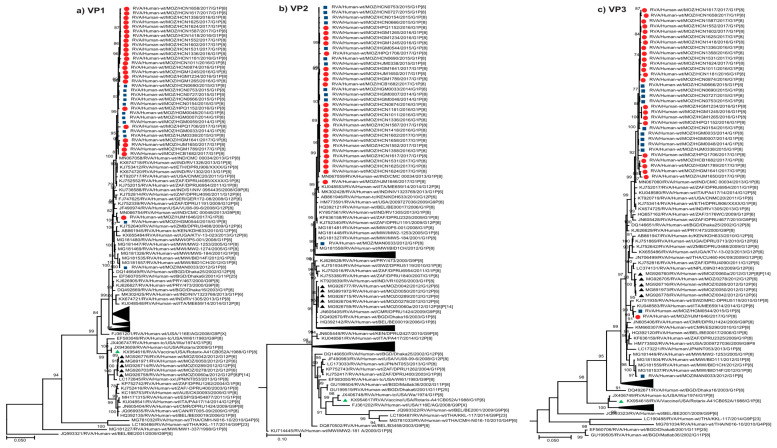
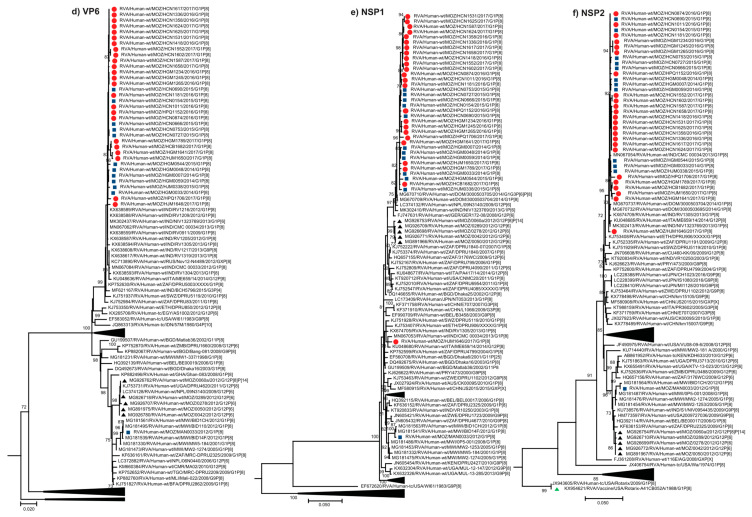
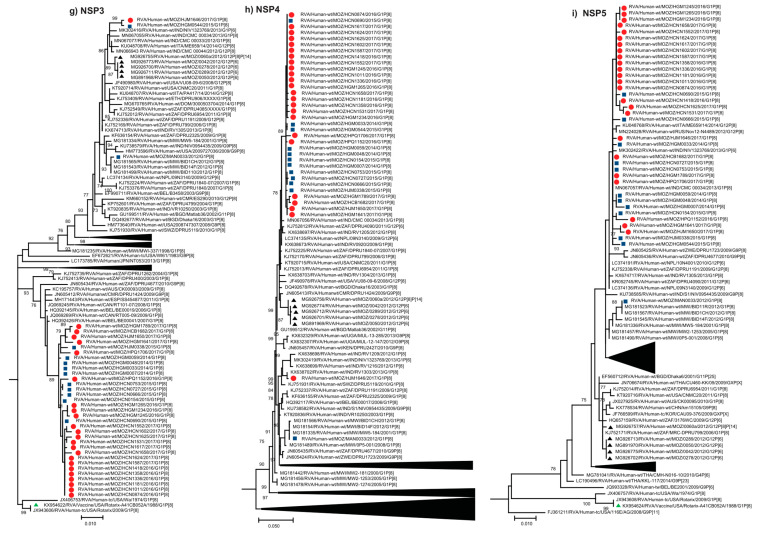
Figure 2.
Phylogenetic trees based on the ORF nucleotide sequences of the (a) VP1, (b) VP2, (c) VP3, (d) VP6, (e) NSP1, (f) NSP2, (g) NSP3, (h) NSP4 and (i) NSP5 genes of G1P[8] strains circulating in Mozambique and global strains obtained from GenBank. The trees were constructed based on the maximum likelihood method implemented in MEGA X [29], using the best-fit nucleotide substitution model General Time Reversible (GTR+G+I) for VP3, GTR+G for VP2, NSP2 and NSP3, Hasegawa Kishino Yano (HKY+G+I) for VP6 and NSP1, HKY+G for VP1, NSP4 and NSP5/6, determined by JModelTest [30]. Bootstrap values (1000 replicates) ≥70% are shown with DS-1 serving as an out-group (not shown in the final tree). Scale bar indicates genetic distance expressed as the number of nucleotide substitutions per site. Pre-vaccine Mozambican strains are indicated by blue squares, post-vaccine by red circles, the Rotarix® vaccine strain by a green triangle and Mozambican strains from a previous study [19] are indicated by black triangles.