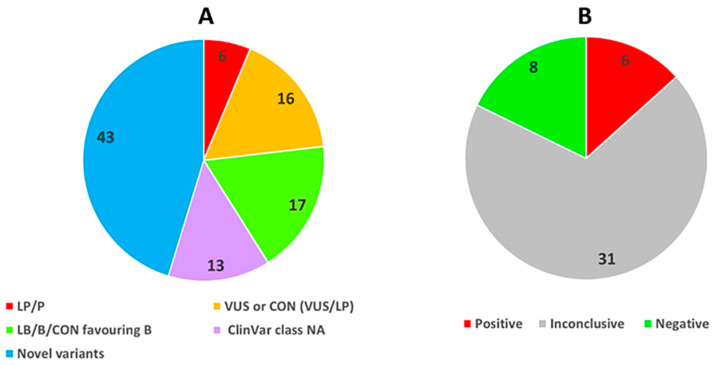
Figure 1.
Percentage distribution of rare variants (AF < 0.001) and detection rates. (A). Type and distribution of variants according to ClinVar classification; novel variants refers to sequence variants not previously published nor reported in online variant databases; mutations within all the others groups were previously published or reported in specific databases. (B). Results of genetic testing within the entire HCM cohort broken down by category. Positive: all cases with a variant classified as LP/P by ClinVar; negative: no rare variant identified or only B/LB variants according to ClinVar; inconclusive: cases with one (or combination) of the following type of variants: variants categorized as VUS, variants for which conflicting interpretations of pathogenicity exists (either VUS/LP or VUS/B/LB), variants without ClinVar classification, and all novel variants irrespective of VarSome classification. AF allele frequency; B benign; CON variant with conflicting interpretations of pathogenicity; LB likely benign; LP likely pathogenic; NA data not available; P pathogenic; VUS variant of uncertain significance.